| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 8742 |
Name | TNFSF12 |
Synonymous | tumor necrosis factor (ligand) superfamily, member 12;TNFSF12;tumor necrosis factor (ligand) superfamily, member 12 |
Definition | APO3 ligand|APO3/DR3 ligand|TNF-related WEAK inducer of apoptosis|tumor necrosis factor ligand superfamily member 12 |
Position | 17p13 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
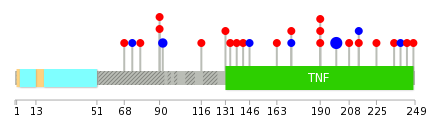 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
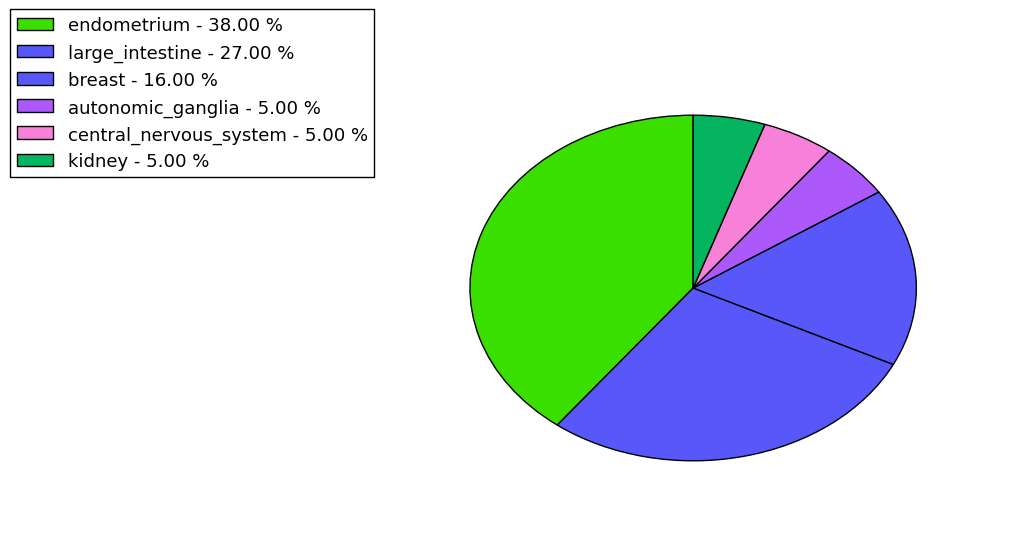 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.06. |
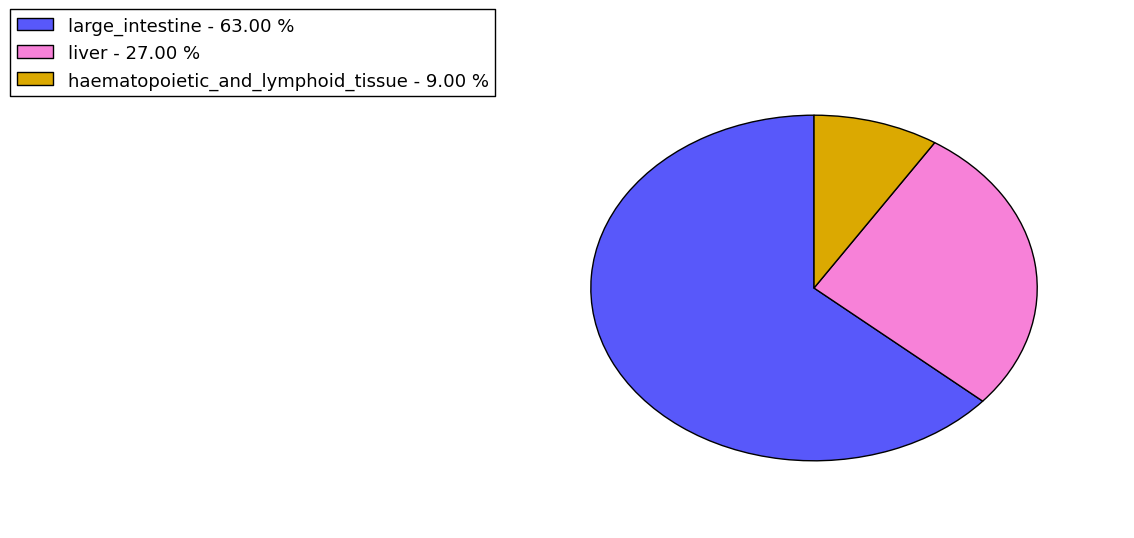
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.268C>T; p.R90W; 17:7550180-7550180 |
breast | carcinoma | Substitution - Missense |
c.414C>G; p.I138M; 17:7556818-7556818 |
breast | carcinoma | Substitution - Missense |
c.269G>A; p.R90Q; 17:7550181-7550181 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.234C>A; p.S78R; 17:7550146-7550146 |
autonomic_ganglia | neuroblastoma | Substitution - Missense |
c.569G>C; p.R190P; 17:7557169-7557169 |
prostate | carcinoma; adenocarcinoma | Substitution - Missense |
c.568C>T; p.R190C; 17:7557168-7557168 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.374-5T>C; p.?; 17:7556773-7556773 |
ovary | other; neoplasm | Unknown |
c.392G>A; p.S131N; 17:7556796-7556796 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.720C>A; p.L240L; 17:7557320-7557320 |
urinary_tract; bladder | carcinoma | Substitution - coding silent |
c.674G>A; p.R225Q; 17:7557274-7557274 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.438C>T; p.Y146Y; 17:7556842-7556842 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.401A>G; p.E134G; 17:7556805-7556805 |
breast | carcinoma | Substitution - Missense |
c.707C>T; p.A236V; 17:7557307-7557307 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.425G>A; p.S142N; 17:7556829-7556829 |
skin | malignant_melanoma | Substitution - Missense |
c.289A>T; p.K97*; 17:7550804-7550804 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.743T>C; p.V248A; 17:7557343-7557343 |
prostate | carcinoma | Substitution - Missense |
c.347G>A; p.R116Q; 17:7550952-7550952 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.219C>T; p.P73P; 17:7550131-7550131 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.488T>C; p.L163P; 17:7556892-7556892 |
large_intestine; colon | carcinoma | Substitution - Missense |
c.640G>A; p.G214S; 17:7557240-7557240 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.730G>A; p.G244R; 17:7557330-7557330 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.516G>A; p.G172G; 17:7557116-7557116 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - coding silent |
c.600G>C; p.A200A; 17:7557200-7557200 |
thyroid | other; neoplasm | Substitution - coding silent |
c.569G>A; p.R190H; 17:7557169-7557169 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.623G>A; p.R208H; 17:7557223-7557223 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.697C>A; p.H233N; 17:7557297-7557297 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.514G>A; p.G172R; 17:7557114-7557114 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.276C>A; p.R92R; 17:7550188-7550188 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.276C>A; p.R92R; 17:7550188-7550188 |
thyroid | other; neoplasm | Substitution - coding silent |
c.202C>G; p.P68A; 17:7549516-7549516 |
central_nervous_system; brain | primitive_neuroectodermal_tumour-medulloblastoma | Substitution - Missense |