| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 9021 |
Name | SOCS3 |
Synonymous | suppressor of cytokine signaling 3;SOCS3;suppressor of cytokine signaling 3 |
Definition | STAT-induced STAT inhibitor 3|cytokine-inducible SH2 protein 3 |
Position | 17q25.3 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
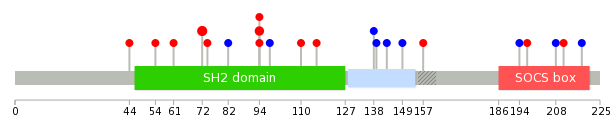 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.08. |
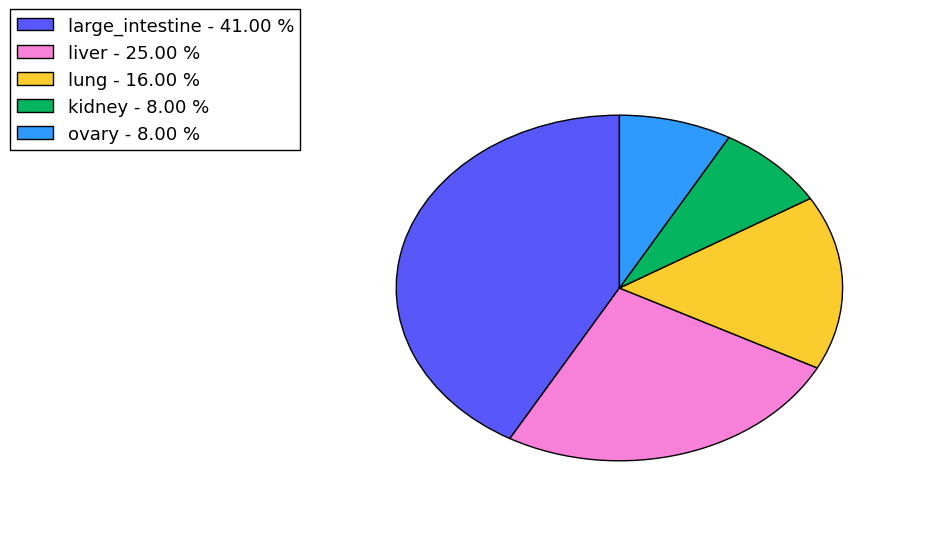
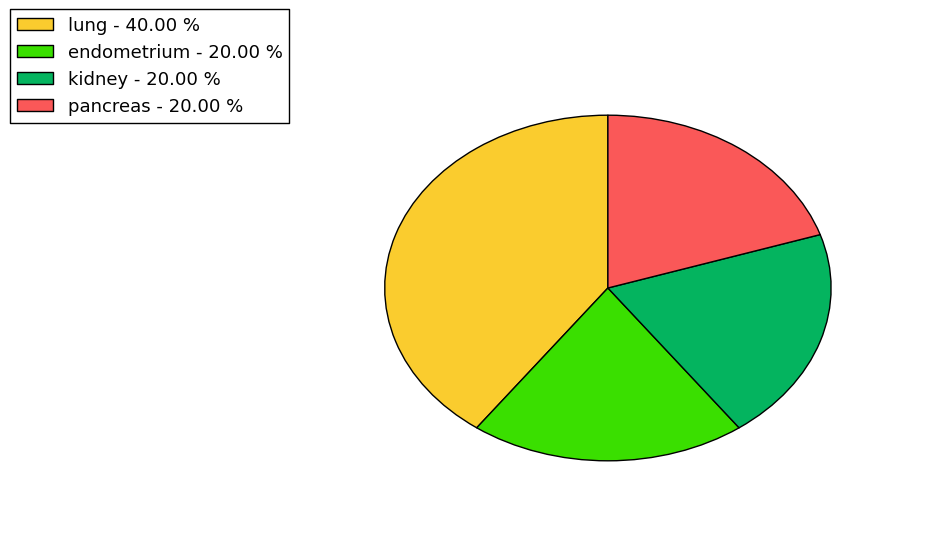
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.318C>T; p.S106S; 17:78358778-78358778 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.92A>G; p.Y31C; 17:78359004-78359004 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.211C>T; p.R71C; 17:78358885-78358885 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.429C>T; p.P143P; 17:78358667-78358667 |
skin | malignant_melanoma | Substitution - coding silent |
c.246C>T; p.L82L; 17:78358850-78358850 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.221C>T; p.S74L; 17:78358875-78358875 |
skin | malignant_melanoma | Substitution - Missense |
c.133G>T; p.G45C; 17:78358963-78358963 |
stomach | carcinoma; intestinal_adenocarcinoma | Substitution - Missense |
c.160G>A; p.G54S; 17:78358936-78358936 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.417A>T; p.P139P; 17:78358679-78358679 |
skin | malignant_melanoma | Substitution - coding silent |
c.212G>A; p.R71H; 17:78358884-78358884 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; peripheral_T_cell_lymphoma_unspecified | Substitution - Missense |
c.624C>T; p.T208T; 17:78358472-78358472 |
skin | malignant_melanoma | Substitution - coding silent |
c.346C>T; p.R116C; 17:78358750-78358750 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.183T>G; p.S61R; 17:78358913-78358913 |
kidney | other; neoplasm | Substitution - Missense |
c.280C>T; p.R94C; 17:78358816-78358816 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.280C>T; p.R94C; 17:78358816-78358816 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.507C>T; p.S169S; 17:78358589-78358589 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.589G>A; p.V197I; 17:78358507-78358507 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.294G>A; p.E98E; 17:78358802-78358802 |
skin | malignant_melanoma | Substitution - coding silent |
c.328A>G; p.S110G; 17:78358768-78358768 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.634G>A; p.G212R; 17:78358462-78358462 |
stomach | carcinoma; diffuse_adenocarcinoma | Substitution - Missense |
c.214G>T; p.D72Y; 17:78358882-78358882 |
liver | carcinoma | Substitution - Missense |
c.214G>T; p.D72Y; 17:78358882-78358882 |
liver | carcinoma | Substitution - Missense |
c.214G>T; p.D72Y; 17:78358882-78358882 |
liver | carcinoma | Substitution - Missense |
c.624C>A; p.T208T; 17:78358472-78358472 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.299delG; p.G100fs*79; 17:78358797-78358797 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.299delG; p.G100fs*79; 17:78358797-78358797 |
large_intestine; caecum | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.299delG; p.G100fs*79; 17:78358797-78358797 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.632C>T; p.P211L; 17:78358464-78358464 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.414G>A; p.S138S; 17:78358682-78358682 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.131G>A; p.S44N; 17:78358965-78358965 |
skin; arm | malignant_melanoma | Substitution - Missense |
c.281G>T; p.R94L; 17:78358815-78358815 |
ovary | carcinoma; clear_cell_carcinoma | Substitution - Missense |
c.347G>A; p.R116H; 17:78358749-78358749 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.218G>A; p.S73N; 17:78358878-78358878 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.654G>A; p.L218L; 17:78358442-78358442 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.582G>A; p.R194R; 17:78358514-78358514 |
pancreas | carcinoma; adenocarcinoma | Substitution - coding silent |
c.447G>A; p.E149E; 17:78358649-78358649 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.281G>A; p.R94H; 17:78358815-78358815 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.469C>T; p.P157S; 17:78358627-78358627 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |