| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 3579 |
Name | CXCR2 |
Synonymous | CD182|CDw128b|CMKAR2|IL8R2|IL8RA|IL8RB;chemokine (C-X-C motif) receptor 2;CXCR2;chemokine (C-X-C motif) receptor 2 |
Definition | C-X-C chemokine receptor type 2|CXC-R2|CXCR-2|CXCR2 gene for IL8 receptor type B|GRO/MGSA receptor|IL-8 receptor type 2|IL-8R B|chemokine (CXC) receptor 2|high affinity interleukin-8 receptor B|interleukin 8 receptor B|interleukin 8 receptor type 2|interl |
Position | 2q35 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
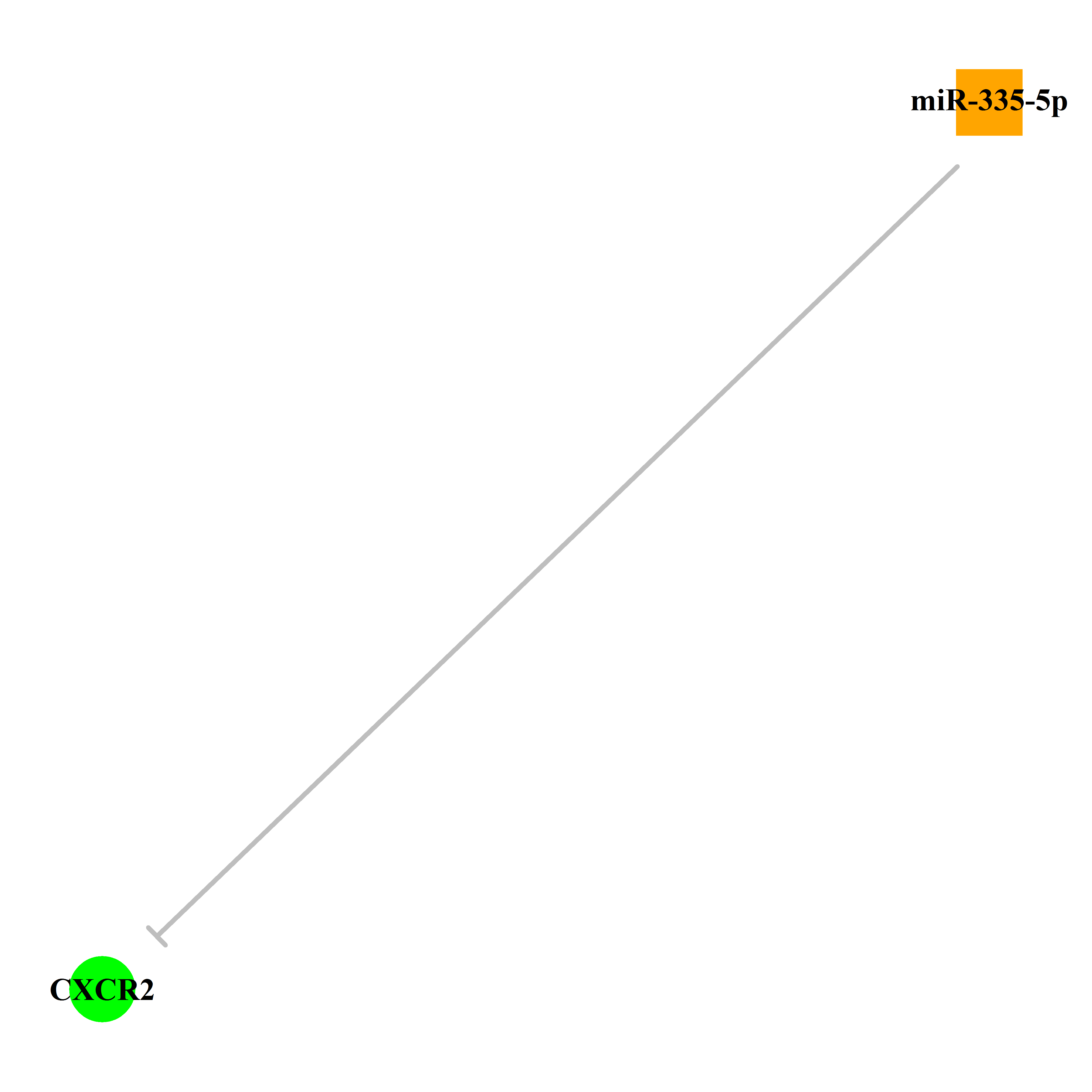 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
IL-8(h) + CXCR2(h) <==> IL-8(h):CXCR2(h) (binding) | CXCR2(h) + PR65(h) <==> CXCR2(h):PR65(h) (binding) | GROalpha(h) + CXCR2(h) <==> GROalpha(h):CXCR2(h) (binding) | NAP-2(h) + CXCR2(h) <==> NAP-2(h):CXCR2(h) (binding) | ENA-78(h) + CXCR2(h) <==> ENA-78(h):CXCR2(h) (binding) | CXCR2(h) + alpha-adaptin(h) <==> CXCR2(h):alpha-adaptin(h) (binding) | CXCR2(h) + beta-adaptin(h) <==> CXCR2(h):beta-adaptin(h) (binding) | CXCR2(h) --> G-alpha-14(m) (activation) | CXCR2(h) --> G-alpha-15(m) (activation) | CXCR2(h) --> G-alpha-15(h) (activation) | CXCR2(h) + Hip(h) <==> CXCR2(h):Hip(h) (binding) | CXCR2(h) + Hip(r) <==> CXCR2(h):Hip(r) (binding) | CXCR2(h) + Rab7(h) <==> CXCR2(h):Rab7(h) (binding) | CXCR2(h) + RAB11(m) <==> CXCR2(h):RAB11(m) (binding) | CXCR2(h) + Rab5A(h) <==> CXCR2(h):Rab5A(h) (binding) | CXCR2(h) + RAB11(h) <==> CXCR2(h):RAB11(h) (binding) | 2 CXCR2(h) <==> (CXCR2(h))2 (binding) | GluR1(h) + CXCR2(h) <==> GluR1(h):CXCR2(h) (binding) | CXCR2(h) + beta-arrestin1(v.s.) <==> CXCR2(h):beta-arrestin1(v.s.) (binding) | CXCR2(h) + G-alpha-i2(h) <==> CXCR2(h):G-alpha-i2(h) (binding) | CXCR2(h) + RGS12(h) <==> CXCR2(h):RGS12(h) (binding) | CXCR2(h){p} --PP2A(h)--> CXCR2(h) + p (dephosphorylation) | IL8RB(h) --> CXCR2(h) (expression). | NAP-2(m.s.) --/ CXCR2(h) (decrease of abundance) | CTAP-III(m.s.) --/ CXCR2(h) (decrease of abundance) | NAP-2-CTF(m.s.) --/ CXCR2(h) (decrease of abundance) | CXCR2(h) + RGS12-isoform1(r) <==> CXCR2(h):RGS12-isoform1(r) (binding) | IL-8(h) + CXCR2(h) <==> IL-8(h):CXCR2(h) (binding) | CXCR2(h) + ATP --> CXCR2(h){p} + ADP (phosphorylation) | CXCR2(h) --> ERK1(r){p} (increase of phosphorylation) | CXCR2(h) --> ERK2(r){p} (increase of phosphorylation) | IL8RB(h) --> CXCR2(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
22 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 347 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 351 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 352 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 353 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 347 | Phosphoserine | 9079638 | Phospho.ELM 6.0 | 351 | Phosphoserine | 9079638 | Phospho.ELM 6.0 | 352 | Phosphoserine | 9079638 | Phospho.ELM 6.0 | 353 | Phosphoserine | 9079638 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
There is no record for CXCR2 | ||