| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 5770 |
Name | PTPN1 |
Synonymous | PTP1B;protein tyrosine phosphatase, non-receptor type 1;PTPN1;protein tyrosine phosphatase, non-receptor type 1 |
Definition | protein tyrosine phosphatase, placental|protein-tyrosine phosphatase 1B|tyrosine-protein phosphatase non-receptor type 1 |
Position | 20q13.1-q13.2 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
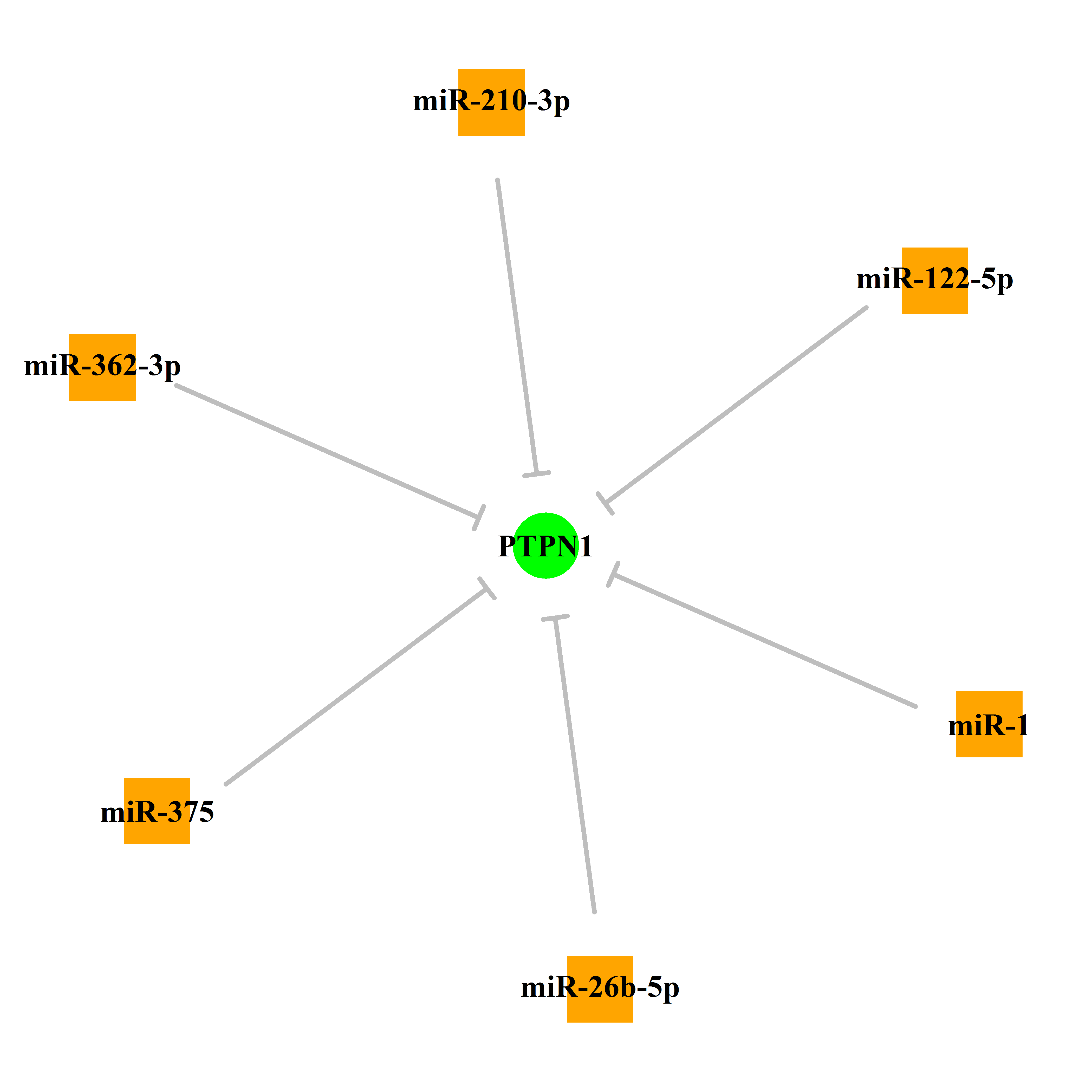 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
PTP1B(h) + Cas(h) <==> PTP1B(h):Cas(h) (binding) | PTP1B(h) --calpain-1(h)--> protein remnants + PTP1B-p42(h) (cleavage) | PTP1B(h) --calpain-2(h)--> protein remnants + PTP1B-p42(h) (cleavage) | Trpv6(r) + PTP1B(h) <==> Trpv6(r):PTP1B(h) (binding) | PTP1B(h) + ATP --Cdk1(m.s.)--> PTP1B(h){pS386} + ADP (phosphorylation) | PTP1B(h) + ATP --PKC(v.s.)--> PTP1B(h){pS378} + ADP (phosphorylation) | PTP1B(h) + ATP --AKT-1(m)--> PTP1B(h){pS50} + ADP (phosphorylation) | PTP1B(h) + InsR(h){pY} <==> PTP1B(h):InsR(h){pY} (binding) | PTP1B(h) + LAT(h){p} --> PTP1B(h):LAT(h) + p (dephosphorylation; binding) | PTP1B(h) --/ IGF1(h) (transrepression) | PTP1B(h) --/ IGF-1(h) (decrease of abundance) | PTP1B(h) --> STAT5(h){pY} (decrease of phosphorylation) | H2O2 --/ PTP1B(h) (inhibition) | superoxide --/ PTP1B(h) (inhibition) | PTPN1(h) --> PTP1B(h) (expression). | Crk-II(m.s.){pY221} --PTP1B(h)--> Crk-II(m.s.) + p (dephosphorylation) | Trpv6(r){pY} --PTP1B(h)--> Trpv6(r) + p (dephosphorylation) | Src(h){pY419}{pY530} --PTP1B(h)--> Src(h) + 2 p (dephosphorylation) | Src(h){pY530} --PTP1B(h)--> Src(h) + p (dephosphorylation) | InsR(h){pY} --PTP1B(h)--> InsR(h) + p (dephosphorylation) | IRS-1(h){pY} --PTP1B(h)--> IRS-1(h) + p (dephosphorylation) | Lck(m){pY} --PTP1B(h)--> Lck(m) + p (dephosphorylation) | LYZ(m.s.){pY} --PTP1B(h)--> p + LYZ(m.s.) (dephosphorylation) | Myosin-P-light chain(m.s.){pY} --PTP1B(h)--> p + Myosin-P-light chain(m.s.) (dephosphorylation) | Albumin(b){pY} --PTP1B(h)--> p + Albumin(b) (dephosphorylation) | MBP(m.s.){pY} --PTP1B(h)--> p + MBP(m.s.) (dephosphorylation) | PTPN1(h) --> PTP1B(h) (expression). | GHR(rb){pY} --PTP1B(h)--> GHR(rb) + p (dephosphorylation) | STAT5(h){pY} --PTP1B(h)--> STAT5(h) + p (dephosphorylation) | STAT3(h){pY} --PTP1B(h)--> STAT3(h) + p (dephosphorylation) |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
1 | N-acetylmethionine. | Swiss-Prot 53.0 | 20 | Phosphotyrosine. | Swiss-Prot 53.0 | 50 | Phosphoserine (by PKB/AKT1). | Swiss-Prot 53.0 | 295 | Phosphoserine. | Swiss-Prot 53.0 | 352 | Phosphoserine. | Swiss-Prot 53.0 | 378 | Phosphoserine (by PKC). | Swiss-Prot 53.0 | 386 | Phosphoserine (by CDC2). | Swiss-Prot 53.0 | 393 | Phosphoserine (By similarity). | Swiss-Prot 53.0 | 215 | 4-amino-3-isothiazolidinone serine (Cys-Ser) (in inhibited form). | Swiss-Prot 53.0 | 352 | Phosphoserine | 93259136 | Phospho.ELM 6.0 | 378 | Phosphoserine (PKC alpha;PK) | 93259136;8491187 | Phospho.ELM 6.0 | 66 | Phosphotyrosine (EGFR;InsR) | 11106648;8999839;9355745 | Phospho.ELM 6.0 | 152 | Phosphotyrosine (InsR) | 11106648 | Phospho.ELM 6.0 | 153 | Phosphotyrosine (InsR) | 11106648 | Phospho.ELM 6.0 | 20 | Phosphotyrosine | 15592455 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr20 |
Promoter: 48558798 - 48560798 | Agilent_014791 |
chr20 |
Promoter: 48558798 - 48560798 | GSE27584 |
chr20 |
Promoter: 48558798 - 48560798 | Agilent_014791_44K |