| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 5781 |
Name | PTPN11 |
Synonymous | BPTP3|CFC|NS1|PTP-1D|PTP2C|SH-PTP2|SH-PTP3|SHP2;protein tyrosine phosphatase, non-receptor type 11;PTPN11;protein tyrosine phosphatase, non-receptor type 11 |
Definition | PTP-2C|protein-tyrosine phosphatase 1D|protein-tyrosine phosphatase 2C|tyrosine-protein phosphatase non-receptor type 11 |
Position | 12q24 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
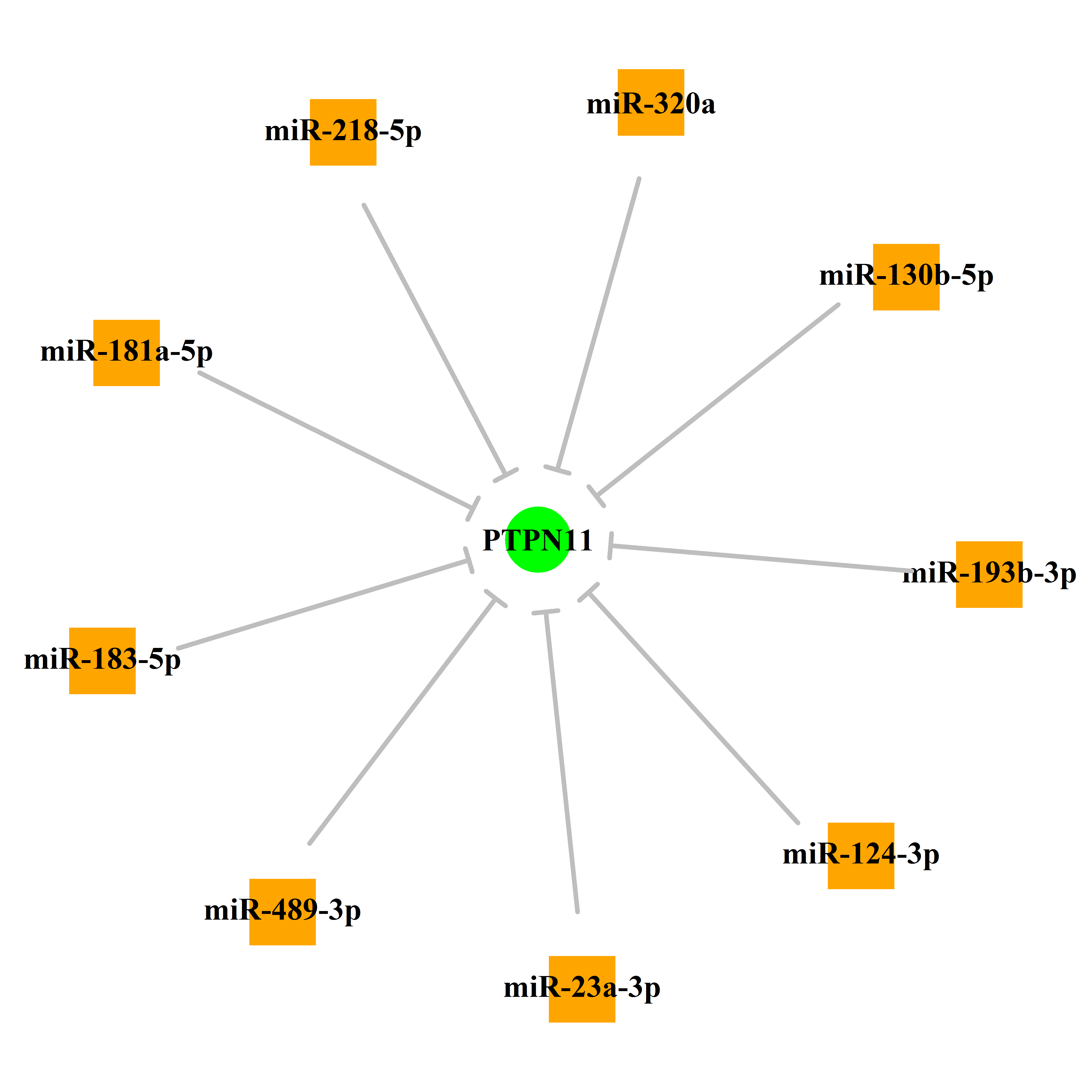 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
ROS(m) + SHP-2(h) <==> ROS(m):SHP-2(h) (binding) | c-Kit(m){pY567} + SHP-2(h) <==> c-Kit(m){pY567}:SHP-2(h) (binding) | SHP-2(h) + MVP(h) <==> SHP-2(h):MVP(h) (binding) | SHP-2(h) + MVP(h){pY} <==> SHP-2(h):MVP(h){pY} (binding) | SHP-2(h) + STAT5A(h) <==> SHP-2(h):STAT5A(h) (binding) | SHP-2(h) --> STAT5A(h){pY694} (increase of phosphorylation) | SHP-2(h) --> SHP-2(h) (translocation) | SHP-2(h) + ATP --> SHP-2(h){pY} + ADP (phosphorylation) | SHP-2(h) + NKAT2(h){pY282}{pY312} <==> SHP-2(h):NKAT2(h){pY282}{pY312} (binding) | SHP-2(h) + kir3dl2(h) <==> SHP-2(h):kir3dl2(h) (binding) | SHP-2(h) + NKAT1(h) <==> SHP-2(h):NKAT1(h) (binding) | SHP-2(h) + NKAT3(h) <==> SHP-2(h):NKAT3(h) (binding) | SHP-2(h) + KIR103AS(h) <==> SHP-2(h):KIR103AS(h) (binding) | MEKK4(h) + SHP-2(h) <==> MEKK4(h):SHP-2(h) (binding) | SHP-2(h) + KLRD1(h):NKG2-A(h){pY} <==> SHP-2(h):KLRD1(h):NKG2-A(h){pY} (binding) | SHP-2(h) + Gab-1(h){pY} <==> SHP-2(h):Gab-1(h){pY} (binding) | paxillin(h) + SHP-2(h) <==> paxillin(h):SHP-2(h) (binding) | SHP-2(h) + Cas(h) <==> SHP-2(h):Cas(h) (binding) | ErbB1-p170(h){pY1016} + SHP-2(h) <==> ErbB1-p170(h){pY1016}:SHP-2(h) (binding) | ErbB2(h){pY1139} + SHP-2(h) <==> ErbB2(h){pY1139}:SHP-2(h) (binding) | ErbB4(h){pY1162} + SHP-2(h) <==> ErbB4(h){pY1162}:SHP-2(h) (binding) | FRS2(h) + SHP-2(h) <==> FRS2(h):SHP-2(h) (binding) | SHP-2(h) + ErbB1(h){pY} <==> SHP-2(h):ErbB1(h){pY} (binding) | SHP-2(h) + Jak2(h):prlr(m.s.) <==> SHP-2(h):Jak2(h):prlr(m.s.) (binding) | SHP-2(h) + p85(h) <==> SHP-2(h):p85(h) (binding) | SHPS1(m.s.) + SHP-2(h) <==> SHPS1(m.s.):SHP-2(h) (binding) | SHP-2(h) + gp130(h) <==> SHP-2(h):gp130(h) (binding) | SHP-2(h) + ATP --Jak1(h)--> SHP-2(h){p} + ADP (phosphorylation) | SHP-2(h) + Jak1(m.s.) <==> SHP-2(h):Jak1(m.s.) (binding) | SHP-2(h) + Tyk2(m.s.) <==> SHP-2(h):Tyk2(m.s.) (binding) | SHP-2(h) + LAIR(h) <==> SHP-2(h):LAIR(h) (binding) | CD244(m.s.){pY} + SHP-2(h) <==> CD244(m.s.){pY}:SHP-2(h) (binding) | IL-5Rbeta(h) + SHP-2(h) <==> IL-5Rbeta(h):SHP-2(h) (binding) | SHP-2(h) + icam1(m.s.){pY} <==> SHP-2(h):icam1(m.s.){pY} (binding) | SHP-2(h) + IRp60(h) <==> SHP-2(h):IRp60(h) (binding) | SHP-2(h) --/ IGF1(h) (transrepression) | SHP-2(h) --/ IGF-1(h) (decrease of abundance) | SHP-2(h) + ATP --> SHP-2(h){pY580} + ADP (phosphorylation) | Jak1(h) + SHP-2(h) + PKCbeta(h) + RACK1(h) <==> RACK1(h):SHP-2(h):Jak1(h):PKCbeta(h) (binding) | Grb-2(h) + SHP-2(h) <==> Grb-2(h):SHP-2(h) (binding) | gp130(m){pY757} + SHP-2(h) <==> gp130(m){pY757}:SHP-2(h) (binding) | PTPN11(h) --> SHP-2(h) (expression). | SHP-2(h) --> SHP-2(h) (translocation) | GM-CSF(m.s.) --> SHP-2(h) (increase of abundance) | PDGFRbeta(v.s.){p} --SHP-2(h)--> PDGFRbeta(v.s.) + p (dephosphorylation) | MVP(h){pY} --SHP-2(h)--> MVP(h) + p (dephosphorylation) | MEKK4(h){pY} --SHP-2(h)--> p + MEKK4(h) (dephosphorylation) | Pyk2(h){pY} --SHP-2(h)--> p + Pyk2(h) (dephosphorylation) | Src(h){pY} --SHP-2(h)--> p + Src(h) (dephosphorylation) | GHR(rb){pY} --SHP-2(h)--> GHR(rb) + p (dephosphorylation) | PZR(h){p} --SHP-2(h)--> PZR(h) + p (dephosphorylation) | SHP2-isoform2(h) + ATP --> SHP2-isoform2(h){pY542} + ADP (phosphorylation) | PTPN11(h) --> SHP2-isoform2(h) (expression). | PTPN11(h) --> SHP2-isoform1(h) (expression). | PTPN11(h) --> SHP2-isoform3(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
62 | Phosphotyrosine. | Swiss-Prot 53.0 | 546 | Phosphotyrosine (by PDGFR). | Swiss-Prot 53.0 | 584 | Phosphotyrosine (by PDGFR). | Swiss-Prot 53.0 | 546 | Phosphotyrosine (PDGFR beta;P) | 11684012;8041791;12531430;12721296 | Phospho.ELM 6.0 | 62 | Phosphotyrosine | 15592455 | Phospho.ELM 6.0 | 304 | Phosphotyrosine | 12531430 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr12 |
Promoter: 111339419 - 111341419 | GSE27584 |
chr12 |
Promoter: 111339419 - 111341419 | Agilent_014791_44K |