| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 5934 |
Name | RBL2 |
Synonymous | P130|Rb2;retinoblastoma-like 2;RBL2;retinoblastoma-like 2 |
Definition | 130 kDa retinoblastoma-associated protein|PRB2|RBR-2|retinoblastoma-like protein 2|retinoblastoma-related protein 2 |
Position | 16q12.2 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
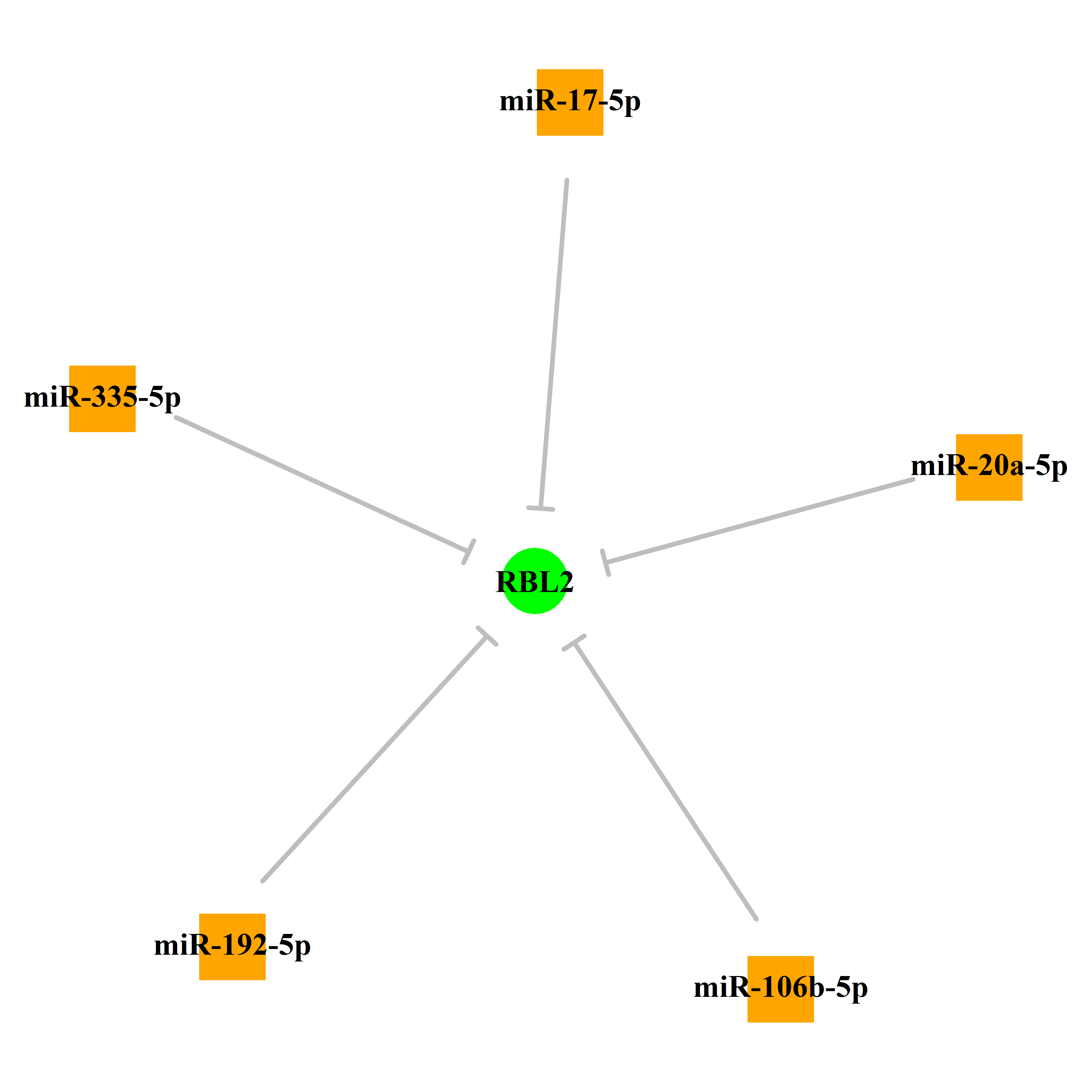 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
p130(h) + cyclinA(h):Cdk2(h) <==> p130(h):cyclinA(h):Cdk2(h) (binding) | p130(h) + cyclinE(h):Cdk2(h) <==> p130(h):cyclinE(h):Cdk2(h) (binding) | E1A(12S)(AD12) + p107(h) + p130(h) + pRb(h) + p300(h) <==> E1A(12S)(AD12):p300(h):pRb(h):p130(h):p107(h) (binding) | E2F-5(h):DP-1(h) + p130(h) <==> E2F-5(h):DP-1(h):p130(h) (binding) | p130(h) + E1A(12S)(AD12) <==> p130(h):E1A(12S)(AD12) (binding) | p130(h) + T-Ag(SV) <==> p130(h):T-Ag(SV) (binding) | p130(h) + cyclinA(h) <==> p130(h):cyclinA(h) (binding) | p130(h) + cyclinD(h) <==> p130(h):cyclinD(h) (binding) | p130(h) + cyclinE(h) <==> p130(h):cyclinE(h) (binding) | E2F-4(h) + p130(h) <==> E2F-4(h):p130(h) (binding) | E2F-4(h) + p130(h) + cyclinA(h):Cdk2(h) <==> E2F-4(h):p130(h):cyclinA(h):Cdk2(h) (binding) | p130(h) + p202(m) <==> p130(h):p202(m) (binding) | E2F-5(h) + p130(h) <==> E2F-5(h):p130(h) (binding). | PMX1(m) + p130(h) <==> PMX1(m):p130(h) (binding). | Alx-4(m) + p130(h) <==> Alx-4(m):p130(h) (binding). | CHX10(m) + p130(h) <==> CHX10(m):p130(h) (binding). | E2F-4(h):DP-1(h) + p130(h) <==> E2F-4(h):DP-1(h):p130(h) (binding) | Raf-1(h) + p130(h) <==> Raf-1(h):p130(h) (binding) | Pax-3(m) + p130(h) <==> Pax-3(m):p130(h) (binding). | p130(h) + E1A(AD) <==> p130(h):E1A(AD) (binding) | p300(h) + p130(h) + E2F-1(h) <==> p300(h):p130(h):E2F-1(h) (binding) | HDAC1(h) + p130(h) + E2F-4(h) <==> HDAC1(h):p130(h):E2F-4(h) (binding) | p130(h) + HDAC1(m.s.) <==> p130(h):HDAC1(m.s.) (binding) | p130(h) + RBBP1(h) <==> p130(h):RBBP1(h) (binding) | E2F-4(h) + p130(h) <==> E2F-4(h):p130(h) (binding) | Cdk2(h) + p130(h) <==> p130(h):Cdk2(h) (binding) | RBL2(h) --> p130(h) (expression). | RBL2(h) --> p130(h) (expression). | RBL2(h) --> RBL2(h) (expression). | hsa-miR-106a --/ RBL2(h) (predicted) | hsa-miR-106b --/ RBL2(h) (predicted) | hsa-miR-20a --/ RBL2(h) (predicted) | hsa-miR-20b --/ RBL2(h) (predicted) |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
413 | Phosphoserine (By similarity). | Swiss-Prot 53.0 | 417 | Phosphothreonine (By similarity). | Swiss-Prot 53.0 | 639 | Phosphoserine. | Swiss-Prot 53.0 | 642 | Phosphothreonine (By similarity). | Swiss-Prot 53.0 | 672 | Phosphoserine. | Swiss-Prot 53.0 | 948 | Phosphoserine (By similarity). | Swiss-Prot 53.0 | 952 | Phosphoserine. | Swiss-Prot 53.0 | 966 | Phosphoserine. | Swiss-Prot 53.0 | 971 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 972 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 973 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 974 | Phosphothreonine (Probable). | Swiss-Prot 53.0 | 981 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 982 | Phosphoserine (Probable). | Swiss-Prot 53.0 | 986 | Phosphothreonine. | Swiss-Prot 53.0 | 639 | Phosphoserine (CDK2) | 11042701;11157749 | Phospho.ELM 6.0 | 952 | Phosphoserine (CDK2) | 11042701;11157749 | Phospho.ELM 6.0 | 966 | Phosphoserine | 11042701 | Phospho.ELM 6.0 | 986 | Phosphothreonine | 11042701 | Phospho.ELM 6.0 | 981 | Phosphoserine | 11042701 | Phospho.ELM 6.0 | 982 | Phosphoserine (CDK2) | 11042701;11157749 | Phospho.ELM 6.0 | 971 | Phosphoserine | 11042701 | Phospho.ELM 6.0 | 972 | Phosphoserine | 11042701 | Phospho.ELM 6.0 | 973 | Phosphoserine (CDK2) | 11042701;11157749 | Phospho.ELM 6.0 | 974 | Phosphothreonine | 11042701 | Phospho.ELM 6.0 | 948 | Phosphoserine | 11042701 | Phospho.ELM 6.0 | 401 | Phosphothreonine (CDK4;CDK6) | 11157749 | Phospho.ELM 6.0 | 672 | Phosphoserine (CDK4;CDK6) | 11157749 | Phospho.ELM 6.0 | 962 | Phosphoserine | 11157749 | Phospho.ELM 6.0 | 1035 | Phosphoserine (CDK4;CDK6) | 11157749 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr16 |
Promoter: 52024401 - 52026401 | Agilent_014791 |
chr16 |
Promoter: 52024401 - 52026401 | GoldenGate_Methylation_Cancer_Panel_I |
chr16 |
Promoter: 52024401 - 52026401 | GSE27584 |
chr16 |
Promoter: 52024401 - 52026401 | Agilent_014791_44K |