| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 8453 |
Name | CUL2 |
Synonymous | -;cullin 2;CUL2;cullin 2 |
Definition | CUL-2|cullin-2 |
Position | 10p11.21 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
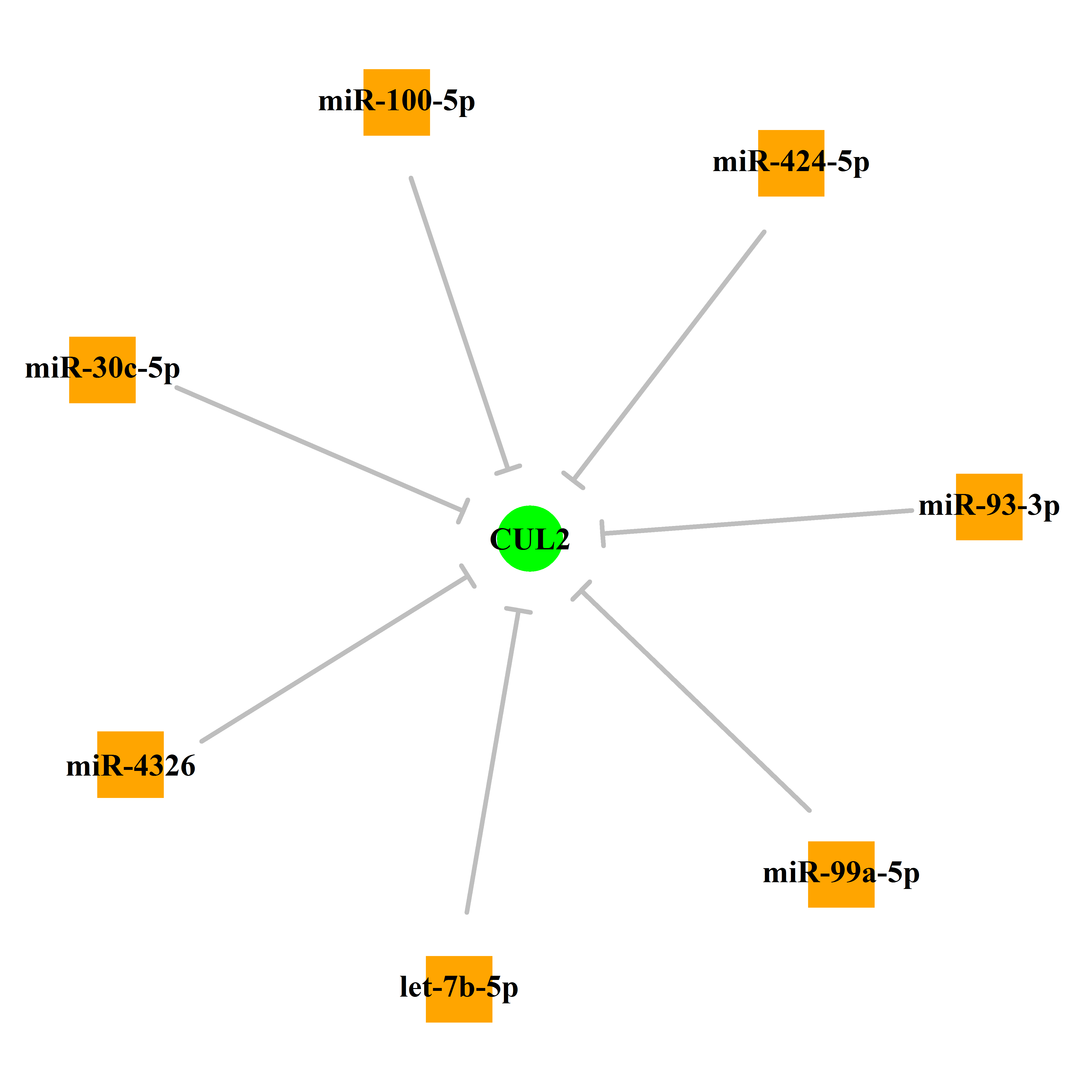 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
Cul-2(h) + Murr1(h) <==> Cul-2(h):Murr1(h) (binding) | VHL(h) + Cul-2(h) <==> VHL(h):Cul-2(h) (binding) | Cul-2(h) + PDI(h) <==> Cul-2(h):PDI(h) (binding) | Cul-2(h) + MO25(h) <==> Cul-2(h):MO25(h) (binding) | TFEA(h) + Cul-2(h) <==> TFEA(h):Cul-2(h) (binding) | Cul-2(h) + TIM13(h) <==> Cul-2(h):TIM13(h) (binding) | Cul-2(h) + STOM(h) <==> Cul-2(h):STOM(h) (binding) | SAP102(h) + Cul-2(h) <==> SAP102(h):Cul-2(h) (binding) | Cul-2(h) + 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3(h) <==> Cul-2(h):6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3(h) (binding) | Cul-2(h) + CMBL(h) <==> Cul-2(h):CMBL(h) (binding) | Cul-2(h) + HC90(h) <==> Cul-2(h):HC90(h) (binding) | Cul-2(h) + KCTD5(h) <==> Cul-2(h):KCTD5(h) (binding) | CUL2(h) --> Cul-2(h) (expression). | CUL2(h) --> Cul-2(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
689 | Glycyl lysine isopeptide (Lys-Gly)(interchain with G-Cter in NEDD8). | Swiss-Prot 53.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr10 |
Promoter: 35418800 - 35420800 | Agilent_014791 |
chr10 |
Promoter: 35418800 - 35420800 | GSE27584 |
chr10 |
Promoter: 35418800 - 35420800 | Agilent_014791_44K |