Gene Page: CDH4
Summary ?
| GeneID | 1002 |
| Symbol | CDH4 |
| Synonyms | CAD4|R-CAD|RCAD |
| Description | cadherin 4 |
| Reference | MIM:603006|HGNC:HGNC:1763|Ensembl:ENSG00000179242|HPRD:04304|Vega:OTTHUMG00000032890 |
| Gene type | protein-coding |
| Map location | 20q13.3 |
| Pascal p-value | 0.087 |
| Fetal beta | 0.98 |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
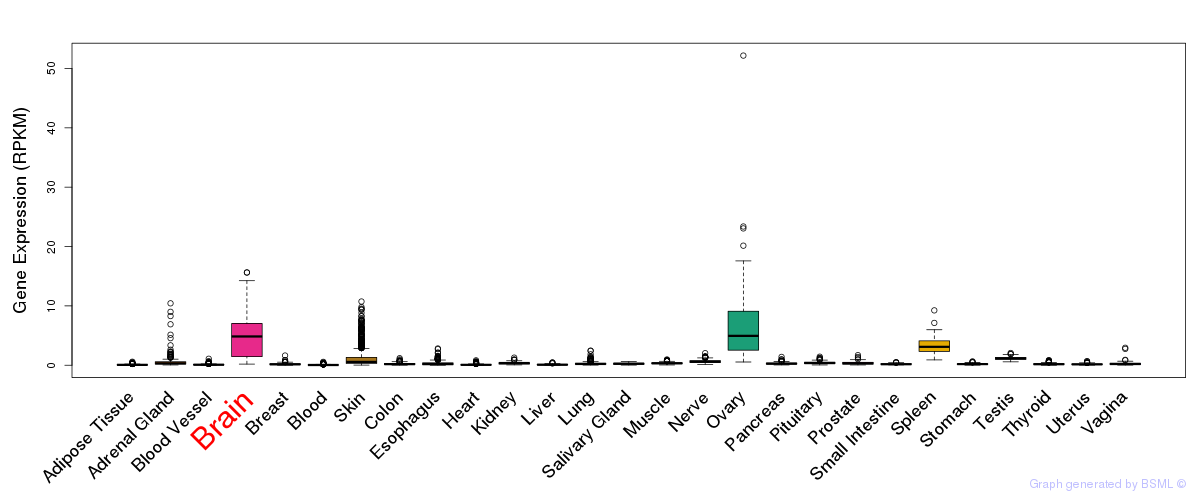
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKRD55 | 0.76 | 0.80 |
| SPATS2L | 0.75 | 0.75 |
| FILIP1 | 0.74 | 0.81 |
| C5orf40 | 0.71 | 0.83 |
| SKAP2 | 0.70 | 0.76 |
| GUCY1B3 | 0.70 | 0.79 |
| FLRT3 | 0.69 | 0.73 |
| TDRD5 | 0.69 | 0.69 |
| GABRA5 | 0.68 | 0.78 |
| OXCT1 | 0.68 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM181B | -0.34 | -0.42 |
| HEBP2 | -0.33 | -0.42 |
| CSRP2 | -0.33 | -0.30 |
| IRF7 | -0.32 | -0.38 |
| METRNL | -0.31 | -0.40 |
| KCNE1 | -0.31 | -0.24 |
| SLA | -0.31 | 0.11 |
| AF347015.18 | -0.31 | -0.16 |
| NHSL1 | -0.30 | -0.23 |
| NEUROD2 | -0.30 | 0.06 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0045773 | positive regulation of axon extension | IEA | axon (GO term level: 15) | - |
| GO:0007155 | cell adhesion | TAS | 2059658 | |
| GO:0007156 | homophilic cell adhesion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| AIGNER ZEB1 TARGETS | 35 | 15 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION DN | 30 | 25 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| NADELLA PRKAR1A TARGETS UP | 9 | 5 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-26 | 243 | 249 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.