Gene Page: TRAP1
Summary ?
| GeneID | 10131 |
| Symbol | TRAP1 |
| Synonyms | HSP 75|HSP75|HSP90L|TRAP-1 |
| Description | TNF receptor-associated protein 1 |
| Reference | MIM:606219|HGNC:HGNC:16264|Ensembl:ENSG00000126602|HPRD:05868|Vega:OTTHUMG00000129427 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.511 |
| Sherlock p-value | 0.595 |
| Fetal beta | -0.316 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Cortex |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0125 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26927547 | 16 | 3766892 | TRAP1 | 4.78E-9 | -0.012 | 2.75E-6 | DMG:Jaffe_2016 |
| cg02634421 | 16 | 3767934 | TRAP1 | 8.52E-8 | -0.008 | 1.97E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
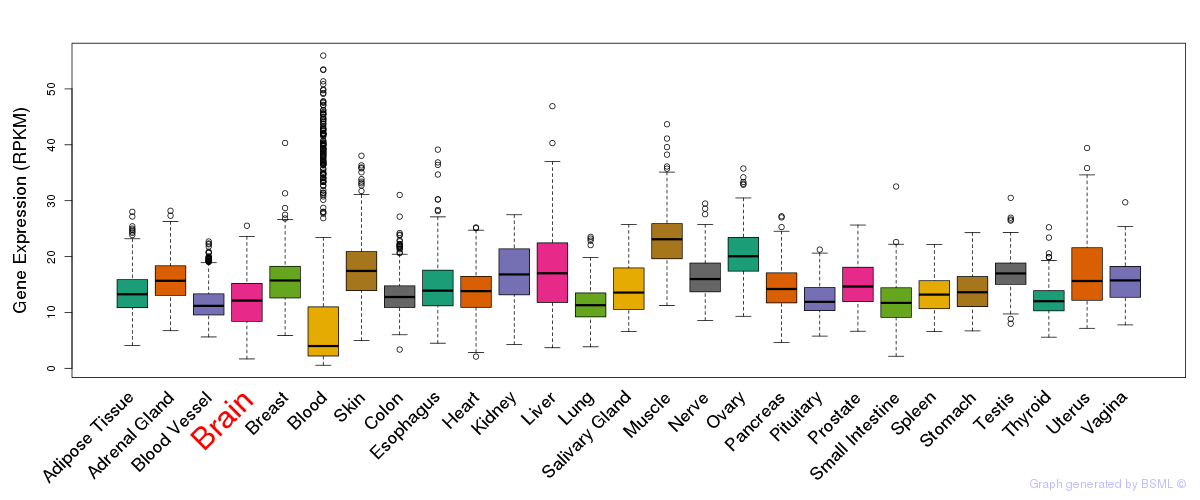
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKRD17 | 0.96 | 0.96 |
| HUWE1 | 0.94 | 0.95 |
| DYRK1A | 0.94 | 0.95 |
| HERC1 | 0.93 | 0.93 |
| VPS39 | 0.93 | 0.93 |
| SEC24B | 0.93 | 0.93 |
| ADD1 | 0.93 | 0.93 |
| KIAA0090 | 0.93 | 0.92 |
| GTF3C1 | 0.93 | 0.92 |
| RPRD2 | 0.93 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.80 | -0.83 |
| MT-CO2 | -0.79 | -0.83 |
| HIGD1B | -0.79 | -0.84 |
| IFI27 | -0.77 | -0.83 |
| FXYD1 | -0.77 | -0.82 |
| AF347015.21 | -0.77 | -0.86 |
| AF347015.8 | -0.75 | -0.80 |
| AF347015.33 | -0.75 | -0.78 |
| MT-CYB | -0.74 | -0.78 |
| CXCL14 | -0.74 | -0.81 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005164 | tumor necrosis factor receptor binding | NAS | 7876093 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0051082 | unfolded protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006457 | protein folding | IEA | - | |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 3 DN | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| COLLER MYC TARGETS UP | 25 | 19 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA UP | 45 | 30 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| CHENG RESPONSE TO NICKEL ACETATE | 45 | 29 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-320 | 53 | 59 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.