Gene Page: ABI2
Summary ?
| GeneID | 10152 |
| Symbol | ABI2 |
| Synonyms | ABI-2|ABI2B|AIP-1|AblBP3|SSH3BP2|argBP1|argBPIA|argBPIB |
| Description | abl-interactor 2 |
| Reference | MIM:606442|HGNC:HGNC:24011|Ensembl:ENSG00000138443|Vega:OTTHUMG00000132879 |
| Gene type | protein-coding |
| Map location | 2q33 |
| Pascal p-value | 0.053 |
| Sherlock p-value | 0.697 |
| Fetal beta | 0.419 |
| DMG | 1 (# studies) |
| Support | STRUCTURAL PLASTICITY CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0043 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05462826 | 2 | 204193327 | ABI2 | -0.021 | 0.34 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
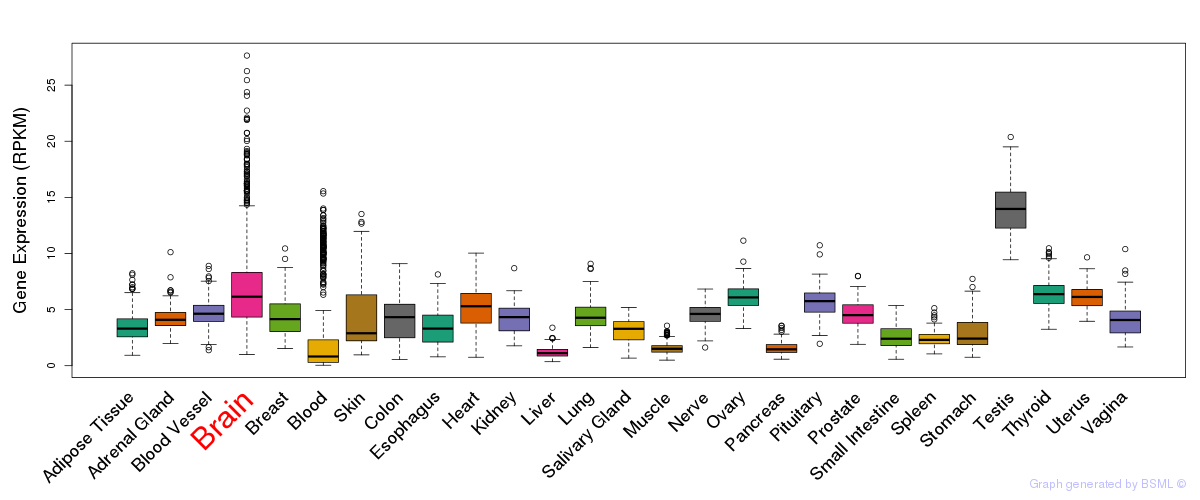
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEATR2 | 0.87 | 0.82 |
| LLGL1 | 0.86 | 0.85 |
| SCRIB | 0.86 | 0.81 |
| RAVER1 | 0.86 | 0.83 |
| PTBP1 | 0.86 | 0.82 |
| ZNF219 | 0.85 | 0.73 |
| ZNF687 | 0.85 | 0.83 |
| AL161668.2 | 0.85 | 0.79 |
| ZNF516 | 0.84 | 0.82 |
| TCF3 | 0.84 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.52 | -0.52 |
| SYCP3 | -0.51 | -0.60 |
| AF347015.31 | -0.48 | -0.53 |
| AF347015.21 | -0.47 | -0.60 |
| C1orf54 | -0.47 | -0.52 |
| CLEC2B | -0.46 | -0.57 |
| CARD16 | -0.46 | -0.56 |
| LY96 | -0.45 | -0.54 |
| RERGL | -0.45 | -0.57 |
| AL050337.1 | -0.44 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0003677 | DNA binding | TAS | 7590236 | |
| GO:0005070 | SH3/SH2 adaptor activity | TAS | 7590236 | |
| GO:0017124 | SH3 domain binding | TAS | 8649853 | |
| GO:0008093 | cytoskeletal adaptor activity | TAS | 12011975 | |
| GO:0019900 | kinase binding | NAS | 8649853 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016358 | dendrite development | IEA | neurite, dendrite (GO term level: 11) | - |
| GO:0007010 | cytoskeleton organization | TAS | 12011975 | |
| GO:0008150 | biological_process | ND | - | |
| GO:0008154 | actin polymerization or depolymerization | NAS | 11516653 | |
| GO:0007611 | learning or memory | IEA | - | |
| GO:0006928 | cell motion | NAS | 11516653 | |
| GO:0018108 | peptidyl-tyrosine phosphorylation | IDA | 17101133 | |
| GO:0016477 | cell migration | IEA | - | |
| GO:0016477 | cell migration | TAS | 12011975 | |
| GO:0043010 | camera-type eye development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030175 | filopodium | NAS | axon (GO term level: 5) | 11516653 |
| GO:0030425 | dendrite | IEA | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0005829 | cytosol | IDA | 17101133 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | NAS | 8649853 | |
| GO:0005737 | cytoplasm | TAS | 8649853 | |
| GO:0030027 | lamellipodium | NAS | 11516653 | |
| GO:0005913 | cell-cell adherens junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 7590236 |
| ADAM19 | FKSG34 | MADDAM | MLTNB | ADAM metallopeptidase domain 19 (meltrin beta) | ADAM19 interacts with ArgBP1. This interaction was modelled on a demonstrated interaction between mouse ADAM19 and human ArgBP1. | BIND | 12463424 |
| CCDC53 | CGI-116 | coiled-coil domain containing 53 | Two-hybrid | BioGRID | 16189514 |
| IFT20 | - | intraflagellar transport 20 homolog (Chlamydomonas) | Two-hybrid | BioGRID | 16189514 |
| KRT15 | CK15 | K15 | K1CO | keratin 15 | Two-hybrid | BioGRID | 16189514 |
| KRT19 | CK19 | K19 | K1CS | MGC15366 | keratin 19 | Two-hybrid | BioGRID | 16189514 |
| KRT20 | CD20 | CK20 | K20 | KRT21 | MGC35423 | keratin 20 | Two-hybrid | BioGRID | 16189514 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| PCM1 | PTC4 | pericentriolar material 1 | Two-hybrid | BioGRID | 16189514 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD,BioGRID | 10858458 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | Two-hybrid | BioGRID | 16189514 |
| VCL | CMD1W | MVCL | vinculin | Two-hybrid | BioGRID | 16189514 |
| VPS37C | FLJ20847 | vacuolar protein sorting 37 homolog C (S. cerevisiae) | - | HPRD | 15509564 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACTINY PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 DN | 45 | 26 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 333 | 339 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-134 | 1003 | 1009 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-144 | 332 | 339 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-186 | 3993 | 3999 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-378 | 326 | 332 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-539 | 350 | 356 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-544 | 361 | 367 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.