Gene Page: PTGES3
Summary ?
| GeneID | 10728 |
| Symbol | PTGES3 |
| Synonyms | P23|TEBP|cPGES |
| Description | prostaglandin E synthase 3 |
| Reference | MIM:607061|HGNC:HGNC:16049|Ensembl:ENSG00000110958|HPRD:06138|Vega:OTTHUMG00000170217 |
| Gene type | protein-coding |
| Map location | 12q13.3|12 |
| Pascal p-value | 0.302 |
| Sherlock p-value | 0.037 |
| Fetal beta | 0.242 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0742 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2403775 | chr8 | 133472361 | PTGES3 | 10728 | 0.15 | trans |
Section II. Transcriptome annotation
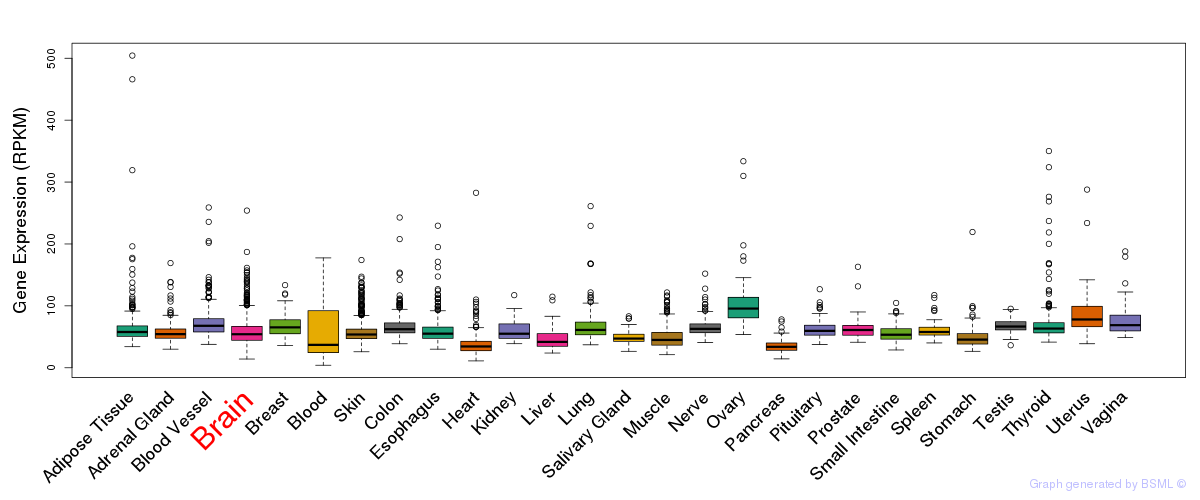
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDYN | 0.60 | 0.63 |
| RARB | 0.58 | 0.63 |
| CDH9 | 0.56 | 0.62 |
| MPP7 | 0.56 | 0.72 |
| KCTD8 | 0.56 | 0.75 |
| VSTM2A | 0.55 | 0.70 |
| AC134043.1 | 0.54 | 0.20 |
| EBF1 | 0.53 | 0.29 |
| FLRT3 | 0.53 | 0.48 |
| HEXB | 0.53 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EMID1 | -0.38 | -0.40 |
| METRNL | -0.35 | -0.48 |
| CBS | -0.33 | -0.38 |
| SLA | -0.30 | -0.16 |
| WDR86 | -0.30 | -0.32 |
| CSRP2 | -0.30 | -0.48 |
| KLHL1 | -0.30 | -0.14 |
| SNX7 | -0.29 | -0.20 |
| TBC1D10A | -0.29 | -0.30 |
| TMEM108 | -0.29 | -0.20 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003720 | telomerase activity | IDA | 12135483 | |
| GO:0016853 | isomerase activity | IEA | - | |
| GO:0050220 | prostaglandin-E synthase activity | IDA | 10922363 | |
| GO:0051082 | unfolded protein binding | IDA | 12077419 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000723 | telomere maintenance | TAS | 12135483 | |
| GO:0001516 | prostaglandin biosynthetic process | IDA | 10922363 | |
| GO:0007165 | signal transduction | TAS | 8114727 | |
| GO:0006633 | fatty acid biosynthetic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000781 | chromosome, telomeric region | IC | 12135483 | |
| GO:0005697 | telomerase holoenzyme complex | IDA | 12135483 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHR | - | aryl hydrocarbon receptor | - | HPRD | 11013261 |11259606 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | - | HPRD | 11879970 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Reconstituted Complex | BioGRID | 11259606 |
| COPG | COPG1 | FLJ21068 | coatomer protein complex, subunit gamma | - | HPRD,BioGRID | 11056392 |
| COPG2 | 2-COP | DKFZp761N09121 | FLJ11781 | coatomer protein complex, subunit gamma 2 | - | HPRD,BioGRID | 11056392 |
| DNAJB1 | HSPF1 | Hdj1 | Hsp40 | Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | - | HPRD | 11809754 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 11447118 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | - | HPRD | 10095121 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Reconstituted Complex | BioGRID | 9222609 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 10207098 |
| FES | FPS | feline sarcoma oncogene | Reconstituted Complex | BioGRID | 9222609 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | Reconstituted Complex | BioGRID | 9222609 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 11812147 |12077419 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | in vivo | BioGRID | 12077419 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 10691735 |
| NR3C2 | MCR | MGC133092 | MLR | MR | nuclear receptor subfamily 3, group C, member 2 | - | HPRD,BioGRID | 10691735 |
| STIP1 | HOP | IEF-SSP-3521 | P60 | STI1 | STI1L | stress-induced-phosphoprotein 1 | - | HPRD | 11809754 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | - | HPRD,BioGRID | 11274138 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 UP | 57 | 34 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| VISALA RESPONSE TO HEAT SHOCK AND AGING DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING UP | 55 | 39 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 538 | 544 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-186 | 853 | 859 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-24* | 262 | 268 | 1A | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.