Gene Page: EHMT2
Summary ?
| GeneID | 10919 |
| Symbol | EHMT2 |
| Synonyms | BAT8|C6orf30|G9A|GAT8|KMT1C|NG36 |
| Description | euchromatic histone-lysine N-methyltransferase 2 |
| Reference | MIM:604599|HGNC:HGNC:14129|Ensembl:ENSG00000204371|HPRD:06854|Vega:OTTHUMG00000031180 |
| Gene type | protein-coding |
| Map location | 6p21.31 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.2 |
| Fetal beta | 0.836 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11203797 | 6 | 31856381 | EHMT2 | 1.824E-4 | 0.481 | 0.034 | DMG:Wockner_2014 |
| cg18424091 | 6 | 31781814 | EHMT2 | 3.26E-6 | 4.451 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10891023 | chr11 | 109738358 | EHMT2 | 10919 | 0.15 | trans |
Section II. Transcriptome annotation
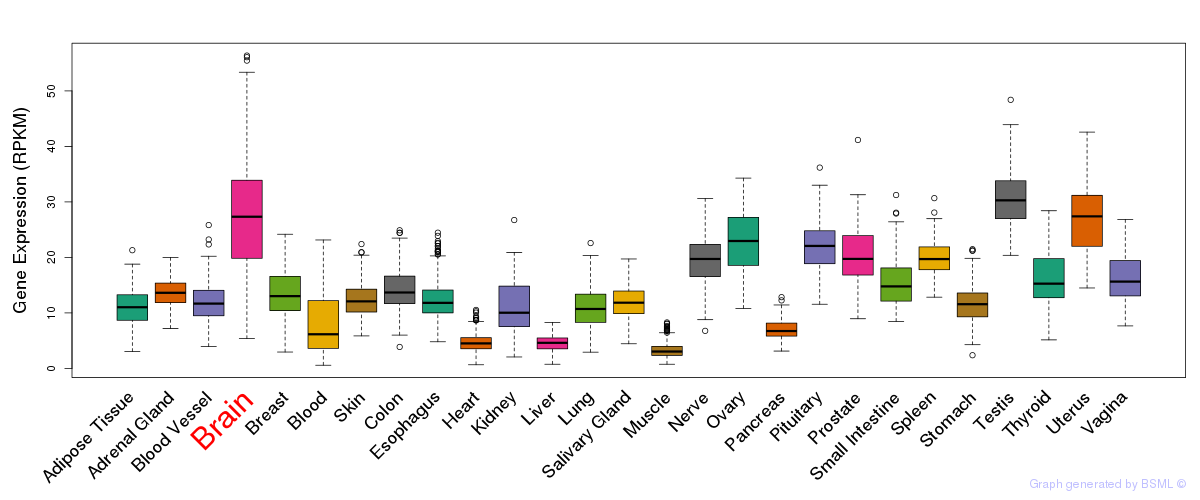
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RCBTB2 | 0.75 | 0.76 |
| CBFB | 0.72 | 0.76 |
| CTPS2 | 0.71 | 0.75 |
| TP53RK | 0.70 | 0.73 |
| ZNF20 | 0.70 | 0.72 |
| SFRS3 | 0.70 | 0.78 |
| MAPRE1 | 0.69 | 0.77 |
| ZNF16 | 0.69 | 0.75 |
| C10orf119 | 0.69 | 0.78 |
| RYK | 0.69 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.54 | -0.75 |
| MT-CO2 | -0.54 | -0.75 |
| HLA-F | -0.53 | -0.68 |
| TINAGL1 | -0.53 | -0.68 |
| FXYD1 | -0.53 | -0.72 |
| AIFM3 | -0.52 | -0.66 |
| MT-CYB | -0.52 | -0.73 |
| AF347015.8 | -0.52 | -0.73 |
| PTH1R | -0.52 | -0.66 |
| ZNF385A | -0.51 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | NAS | 8457211 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008168 | methyltransferase activity | IEA | - | |
| GO:0018024 | histone-lysine N-methyltransferase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007130 | synaptonemal complex assembly | IEA | Synap (GO term level: 11) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0000239 | pachytene | IEA | - | |
| GO:0007286 | spermatid development | IEA | - | |
| GO:0007281 | germ cell development | IEA | - | |
| GO:0008150 | biological_process | ND | - | |
| GO:0009566 | fertilization | IEA | - | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0035265 | organ growth | IEA | - | |
| GO:0051567 | histone H3-K9 methylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARD1A | ARD1 | DXS707 | MGC71248 | TE2 | ARD1 homolog A, N-acetyltransferase (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| BAT2L | DKFZp781F05101 | DKFZp781K12107 | KIAA0515 | LQFBS-1 | MGC10526 | HLA-B associated transcript 2-like | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| C10orf12 | DKFZp564P1916 | FLJ13022 | chromosome 10 open reading frame 12 | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| E4F1 | E4F | MGC99614 | E4F transcription factor 1 | Two-hybrid | BioGRID | 16189514 |
| H3F3B | H3.3B | H3F3A | H3 histone, family 3B (H3.3B) | Two-hybrid | BioGRID | 16189514 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | Two-hybrid | BioGRID | 16189514 |
| KLF12 | AP-2rep | AP2REP | HSPC122 | Kruppel-like factor 12 | Two-hybrid | BioGRID | 16189514 |
| KLF6 | BCD1 | COPEB | CPBP | DKFZp686N0199 | GBF | PAC1 | ST12 | ZF9 | Kruppel-like factor 6 | Two-hybrid | BioGRID | 16189514 |
| LIMS1 | PINCH | PINCH1 | LIM and senescent cell antigen-like domains 1 | Two-hybrid | BioGRID | 16189514 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| PLOD3 | LH3 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | Two-hybrid | BioGRID | 16189514 |
| REST | NRSF | XBR | RE1-silencing transcription factor | NRSF interacts with G9a. | BIND | 15200951 |
| THAP7 | MGC10963 | THAP domain containing 7 | Two-hybrid | BioGRID | 16189514 |
| XAGE2 | GAGED3 | XAGE-2 | X antigen family, member 2 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSINE DEGRADATION | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREEN DN | 25 | 12 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL DN | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| FLOTHO PEDIATRIC ALL THERAPY RESPONSE DN | 29 | 17 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-326 | 177 | 183 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.