Gene Page: CHRNB1
Summary ?
| GeneID | 1140 |
| Symbol | CHRNB1 |
| Synonyms | ACHRB|CHRNB|CMS1D|CMS2A|CMS2C|SCCMS |
| Description | cholinergic receptor nicotinic beta 1 subunit |
| Reference | MIM:100710|HGNC:HGNC:1961|Ensembl:ENSG00000170175|HPRD:01156|Vega:OTTHUMG00000108139 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.071 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22637538 | 17 | 7348327 | CHRNB1 | 3.07E-4 | -0.519 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
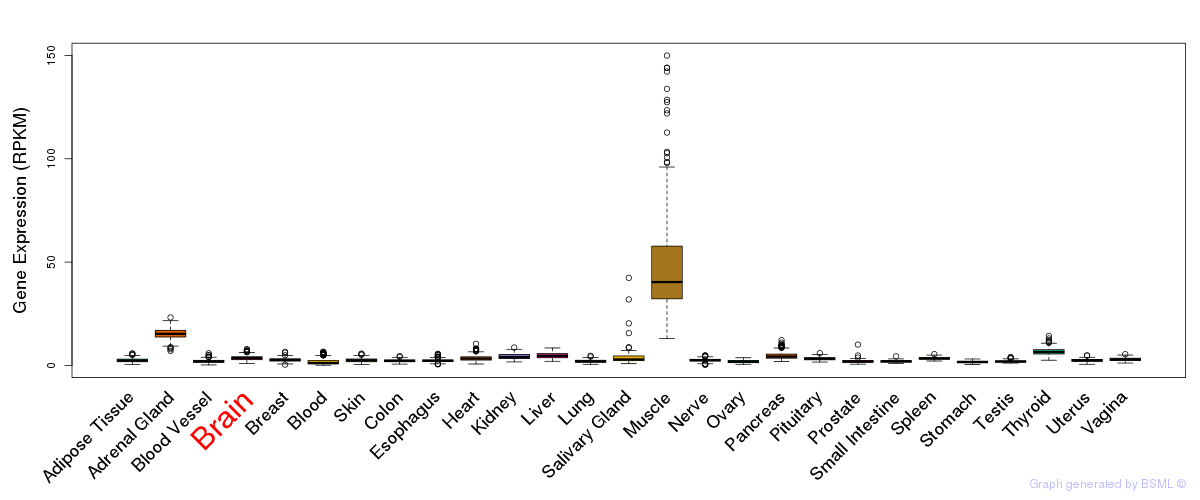
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0015464 | acetylcholine receptor activity | IMP | Neurotransmitter (GO term level: 6) | 8651643 |8872460 |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | ISS | - | |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | ISS | - | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0015267 | channel activity | IMP | 8872460 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007274 | neuromuscular synaptic transmission | IMP | neuron, Synap (GO term level: 7) | 8651643 |
| GO:0007271 | synaptic transmission, cholinergic | IMP | neuron, Cholinergic, Synap, Neurotransmitter (GO term level: 7) | 8872460 |
| GO:0001941 | postsynaptic membrane organization | IMP | Synap (GO term level: 5) | 8651643 |
| GO:0007165 | signal transduction | IMP | 8872460 | |
| GO:0006812 | cation transport | IMP | 8872460 | |
| GO:0006936 | muscle contraction | IMP | 8651643 | |
| GO:0042391 | regulation of membrane potential | ISS | - | |
| GO:0050877 | neurological system process | IMP | 11104662 | |
| GO:0048747 | muscle fiber development | IMP | 8651643 | |
| GO:0035095 | behavioral response to nicotine | IMP | 16874522 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005892 | nicotinic acetylcholine-gated receptor-channel complex | IMP | Neurotransmitter (GO term level: 9) | 8872460 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IMP | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | 8872460 |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACH PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |