Gene Page: CLDN3
Summary ?
| GeneID | 1365 |
| Symbol | CLDN3 |
| Synonyms | C7orf1|CPE-R2|CPETR2|HRVP1|RVP1 |
| Description | claudin 3 |
| Reference | MIM:602910|HGNC:HGNC:2045|Ensembl:ENSG00000165215|HPRD:04219|Vega:OTTHUMG00000023424 |
| Gene type | protein-coding |
| Map location | 7q11.23 |
| Pascal p-value | 0.352 |
| Fetal beta | -0.388 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | CLDN3 | 1365 | 0.1 | trans | ||
| rs2502827 | chr1 | 176044216 | CLDN3 | 1365 | 0.06 | trans | ||
| rs17572651 | chr1 | 218943612 | CLDN3 | 1365 | 0.09 | trans | ||
| rs16829545 | chr2 | 151977407 | CLDN3 | 1365 | 5.92E-10 | trans | ||
| rs7584986 | chr2 | 184111432 | CLDN3 | 1365 | 1.848E-5 | trans | ||
| rs4575988 | chr4 | 19984925 | CLDN3 | 1365 | 0.18 | trans | ||
| rs170776 | chr4 | 173276735 | CLDN3 | 1365 | 0.15 | trans | ||
| rs1396222 | chr4 | 173279496 | CLDN3 | 1365 | 0.16 | trans | ||
| rs335993 | chr4 | 173321789 | CLDN3 | 1365 | 0.13 | trans | ||
| rs335980 | chr4 | 173329784 | CLDN3 | 1365 | 0.17 | trans | ||
| rs17762315 | chr5 | 76807576 | CLDN3 | 1365 | 0.05 | trans | ||
| rs10491487 | chr5 | 80323367 | CLDN3 | 1365 | 0.02 | trans | ||
| rs1368303 | chr5 | 147672388 | CLDN3 | 1365 | 0.01 | trans | ||
| rs6996695 | chr8 | 77540580 | CLDN3 | 1365 | 0.16 | trans | ||
| rs10984951 | chr9 | 123306781 | CLDN3 | 1365 | 0.11 | trans | ||
| rs4837782 | chr9 | 123330812 | CLDN3 | 1365 | 0.07 | trans | ||
| rs10739568 | chr9 | 123331138 | CLDN3 | 1365 | 0.09 | trans | ||
| rs17263352 | chr9 | 124811572 | CLDN3 | 1365 | 0.17 | trans | ||
| rs457696 | chr13 | 32581313 | CLDN3 | 1365 | 0.03 | trans | ||
| rs9566986 | chr13 | 32595026 | CLDN3 | 1365 | 0.07 | trans | ||
| rs16955618 | chr15 | 29937543 | CLDN3 | 1365 | 4.549E-17 | trans | ||
| rs17823633 | chr17 | 68851458 | CLDN3 | 1365 | 0.07 | trans | ||
| rs17681878 | chr18 | 34767223 | CLDN3 | 1365 | 0.17 | trans | ||
| rs749043 | chr20 | 46995700 | CLDN3 | 1365 | 0.03 | trans |
Section II. Transcriptome annotation
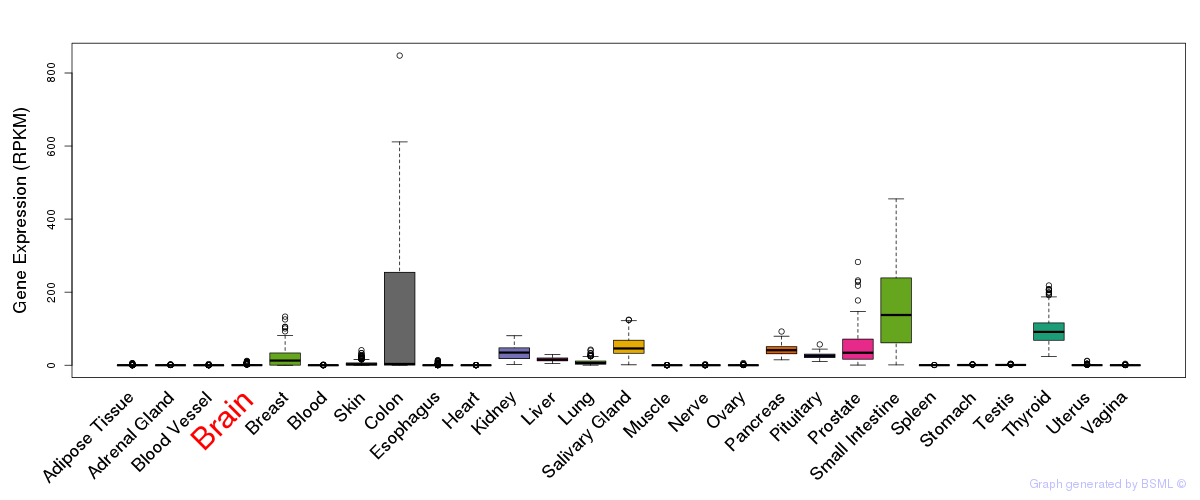
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TPCN1 | 0.90 | 0.87 |
| KANK1 | 0.87 | 0.83 |
| FYCO1 | 0.86 | 0.87 |
| KCNJ10 | 0.85 | 0.82 |
| ACACB | 0.85 | 0.83 |
| ATP1A2 | 0.85 | 0.84 |
| CHDH | 0.84 | 0.80 |
| SASH1 | 0.83 | 0.80 |
| ITPKB | 0.83 | 0.84 |
| TPP1 | 0.83 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.52 | -0.55 |
| POLB | -0.52 | -0.55 |
| RPS3AP47 | -0.49 | -0.63 |
| NR2C2AP | -0.49 | -0.51 |
| STMN1 | -0.49 | -0.48 |
| FRG1 | -0.49 | -0.54 |
| RPL24 | -0.48 | -0.64 |
| COMMD3 | -0.48 | -0.53 |
| ALKBH2 | -0.48 | -0.53 |
| RPL27 | -0.48 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | TAS | 9334247 | |
| GO:0005198 | structural molecule activity | IEA | - | |
| GO:0042802 | identical protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001666 | response to hypoxia | IEP | 15174142 | |
| GO:0016338 | calcium-independent cell-cell adhesion | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | ISS | Brain (GO term level: 10) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9334247 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME TIGHT JUNCTION INTERACTIONS | 29 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| LANDIS BREAST CANCER PROGRESSION UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| CREIGHTON AKT1 SIGNALING VIA MTOR UP | 34 | 22 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |