Gene Page: DTNB
Summary ?
| GeneID | 1838 |
| Symbol | DTNB |
| Synonyms | - |
| Description | dystrobrevin beta |
| Reference | MIM:602415|HGNC:HGNC:3058|Ensembl:ENSG00000138101|HPRD:09091|Vega:OTTHUMG00000152129 |
| Gene type | protein-coding |
| Map location | 2p24 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.337 |
| Fetal beta | -1.237 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2064 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17774001 | 2 | 25600752 | DTNB | 5.48E-4 | 0.588 | 0.049 | DMG:Wockner_2014 |
| cg17692591 | 2 | 25896739 | DTNB | 9.32E-11 | -0.01 | 4.49E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
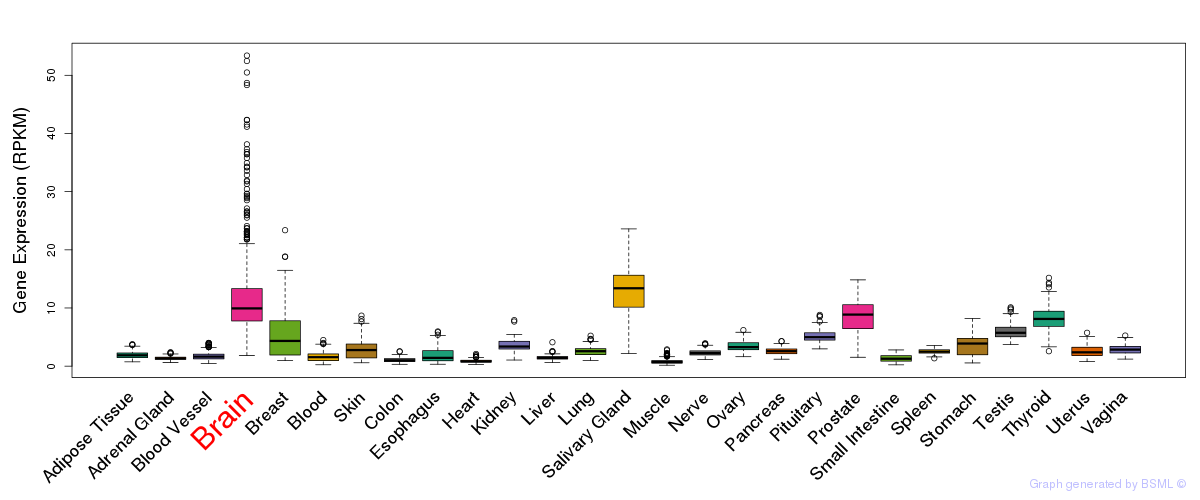
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| JPH4 | 0.91 | 0.92 |
| DMWD | 0.89 | 0.89 |
| FBXO41 | 0.88 | 0.91 |
| REXO1 | 0.87 | 0.89 |
| C11orf81 | 0.87 | 0.88 |
| JPH3 | 0.87 | 0.90 |
| SEMA6B | 0.86 | 0.88 |
| MGAT5B | 0.86 | 0.88 |
| AGAP2 | 0.86 | 0.88 |
| C19orf26 | 0.86 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.56 | -0.47 |
| AL139819.3 | -0.55 | -0.55 |
| GNG11 | -0.54 | -0.53 |
| AP002478.3 | -0.53 | -0.52 |
| C1orf54 | -0.51 | -0.52 |
| MT-CO2 | -0.51 | -0.43 |
| AF347015.31 | -0.50 | -0.43 |
| NOSTRIN | -0.50 | -0.45 |
| AF347015.18 | -0.48 | -0.37 |
| AF347015.8 | -0.48 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | TAS | 9395493 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | Two-hybrid | BioGRID | 16189514 |
| C1orf182 | MGC26877 | RP11-443G18.5 | SSTK-IP | chromosome 1 open reading frame 182 | Two-hybrid | BioGRID | 16189514 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| DMD | BMD | CMD3B | DXS142 | DXS164 | DXS206 | DXS230 | DXS239 | DXS268 | DXS269 | DXS270 | DXS272 | dystrophin | Affinity Capture-Western | BioGRID | 10545507 |
| DTNBP1 | DBND | DKFZp564K192 | FLJ30031 | HPS7 | MGC20210 | My031 | SDY | dystrobrevin binding protein 1 | - | HPRD | 11316798 |
| HMG20A | FLJ10739 | HMGX1 | high-mobility group 20A | Two-hybrid | BioGRID | 16189514 |
| KIF5A | D12S1889 | MY050 | NKHC | SPG10 | kinesin family member 5A | - | HPRD,BioGRID | 14600269 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | Reconstituted Complex | BioGRID | 14600269 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | Two-hybrid | BioGRID | 16189514 |
| PPFIBP2 | Cclp1 | DKFZp781K06126 | MGC42541 | PTPRF interacting protein, binding protein 2 (liprin beta 2) | Two-hybrid | BioGRID | 16189514 |
| PRTFDC1 | FLJ11888 | HHGP | phosphoribosyl transferase domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | Affinity Capture-Western | BioGRID | 10545507 |
| SNTG1 | G1SYN | SYN4 | syntrophin, gamma 1 | - | HPRD | 10747910 |
| SNTG2 | G2SYN | MGC133174 | SYN5 | syntrophin, gamma 2 | - | HPRD | 10747910 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| UTRN | DMDL | DRP | DRP1 | FLJ23678 | utrophin | - | HPRD | 10545507 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER D | 37 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-143 | 284 | 290 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.