| KEGG ERBB SIGNALING PATHWAY
| 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY
| 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY
| 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA
| 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY
| 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY
| 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY
| 20 | 14 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY
| 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY
| 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY
| 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY
| 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION
| 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE
| 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S
| 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS
| 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS
| 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR
| 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MTORC1 MEDIATED SIGNALLING
| 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE
| 71 | 51 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP
| 146 | 86 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN
| 182 | 111 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP
| 74 | 45 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP
| 276 | 187 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP
| 368 | 234 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP
| 204 | 140 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP
| 135 | 82 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN
| 536 | 332 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP
| 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN
| 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP
| 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN
| 153 | 100 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST PRENEOPLASTIC DN
| 55 | 33 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP
| 1142 | 669 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN
| 149 | 93 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL
| 326 | 213 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER
| 140 | 73 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP
| 309 | 199 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE
| 217 | 143 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP
| 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP
| 227 | 137 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN
| 193 | 112 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP
| 479 | 299 | All SZGR 2.0 genes in this pathway |
| OZEN MIR125B1 TARGETS
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS
| 114 | 71 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU UP
| 53 | 37 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON
| 57 | 32 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP
| 142 | 93 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 UP
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS UP
| 33 | 18 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 DN
| 15 | 12 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP
| 206 | 118 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR
| 35 | 22 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN
| 105 | 67 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP
| 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN
| 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN
| 591 | 366 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2
| 118 | 65 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS
| 116 | 63 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP
| 245 | 159 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN
| 122 | 84 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP
| 212 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6
| 456 | 285 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE
| 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13
| 172 | 107 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM
| 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING
| 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT
| 567 | 365 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN
| 374 | 217 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A
| 770 | 480 | All SZGR 2.0 genes in this pathway |
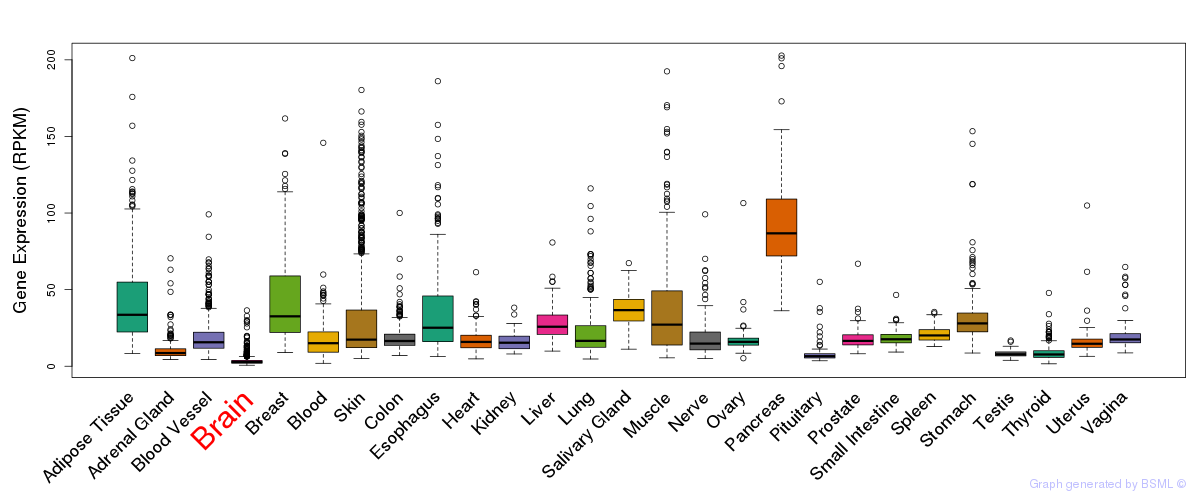
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation