Gene Page: EPHX2
Summary ?
| GeneID | 2053 |
| Symbol | EPHX2 |
| Synonyms | CEH|SEH |
| Description | epoxide hydrolase 2 |
| Reference | MIM:132811|HGNC:HGNC:3402|Ensembl:ENSG00000120915|HPRD:00582|Vega:OTTHUMG00000102115 |
| Gene type | protein-coding |
| Map location | 8p21 |
| Pascal p-value | 8.982E-5 |
| Sherlock p-value | 0.262 |
| Fetal beta | -1.162 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs73229090 | chr8 | 27442127 | AC | 1.952E-8 | intergenic | EPHX2,CLU | dist=39688;dist=12307 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10958899 | chr9 | 10128700 | EPHX2 | 2053 | 0.11 | trans | ||
| rs10958907 | chr9 | 10131580 | EPHX2 | 2053 | 0.09 | trans | ||
| rs1322159 | chr9 | 10132695 | EPHX2 | 2053 | 0.17 | trans | ||
| rs192069 | chrX | 76053555 | EPHX2 | 2053 | 0.07 | trans | ||
| rs16983508 | chrX | 99747940 | EPHX2 | 2053 | 0.12 | trans |
Section II. Transcriptome annotation
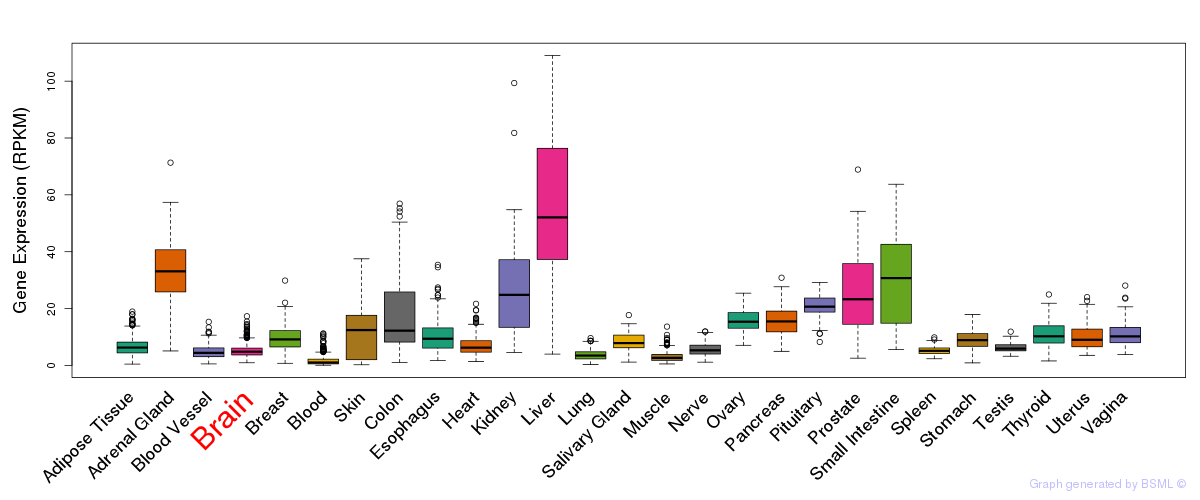
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AAMP | 0.90 | 0.88 |
| GRK6 | 0.89 | 0.88 |
| COBRA1 | 0.89 | 0.88 |
| ATG4B | 0.89 | 0.87 |
| STK25 | 0.89 | 0.86 |
| BBS1 | 0.89 | 0.88 |
| THAP4 | 0.89 | 0.85 |
| RIC8A | 0.89 | 0.88 |
| B4GALT7 | 0.88 | 0.87 |
| TMUB2 | 0.88 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.75 | -0.70 |
| AF347015.21 | -0.75 | -0.74 |
| AF347015.8 | -0.74 | -0.71 |
| AF347015.31 | -0.74 | -0.70 |
| AF347015.27 | -0.73 | -0.72 |
| MT-CYB | -0.72 | -0.69 |
| AF347015.33 | -0.72 | -0.68 |
| AF347015.2 | -0.71 | -0.68 |
| AF347015.15 | -0.70 | -0.67 |
| AF347015.26 | -0.69 | -0.66 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004301 | epoxide hydrolase activity | IDA | 10862610 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | NAS | 10862610 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008217 | regulation of blood pressure | NAS | 10862610 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0009636 | response to toxin | NAS | 10862610 | |
| GO:0017144 | drug metabolic process | NAS | 10862610 | |
| GO:0006805 | xenobiotic metabolic process | NAS | 10862610 | |
| GO:0006800 | oxygen and reactive oxygen species metabolic process | NAS | 10862610 | |
| GO:0006954 | inflammatory response | NAS | 10862610 | |
| GO:0006874 | cellular calcium ion homeostasis | NAS | 10862610 | |
| GO:0019439 | aromatic compound catabolic process | IEA | - | |
| GO:0045909 | positive regulation of vasodilation | NAS | 10862610 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | NAS | 10862610 | |
| GO:0005625 | soluble fraction | NAS | 10862610 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005777 | peroxisome | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARACHIDONIC ACID METABOLISM | 58 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LANDIS BREAST CANCER PROGRESSION DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| YANG MUC2 TARGETS DUODENUM 3MO DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| YANG MUC2 TARGETS DUODENUM 6MO DN | 21 | 12 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| KAPOSI LIVER CANCER MET DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| WANG CLASSIC ADIPOGENIC TARGETS OF PPARG | 26 | 15 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |