Gene Page: B3GAT1
Summary ?
| GeneID | 27087 |
| Symbol | B3GAT1 |
| Synonyms | CD57|GLCATP|GLCUATP|HNK1|LEU7|NK-1|NK1 |
| Description | beta-1,3-glucuronyltransferase 1 |
| Reference | MIM:151290|HGNC:HGNC:921|Ensembl:ENSG00000109956|HPRD:06969|Vega:OTTHUMG00000167180 |
| Gene type | protein-coding |
| Map location | 11q25 |
| Pascal p-value | 3.036E-5 |
| Sherlock p-value | 0.017 |
| Fetal beta | 0.13 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23596233 | 11 | 134281663 | B3GAT1 | 9.89E-8 | -0.009 | 2.2E-5 | DMG:Jaffe_2016 |
| cg19674669 | 11 | 134146910 | B3GAT1 | 0.002 | 4.559 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12528509 | chr6 | 51390611 | B3GAT1 | 27087 | 0.08 | trans | ||
| rs12528978 | chr6 | 51391613 | B3GAT1 | 27087 | 0.17 | trans |
Section II. Transcriptome annotation
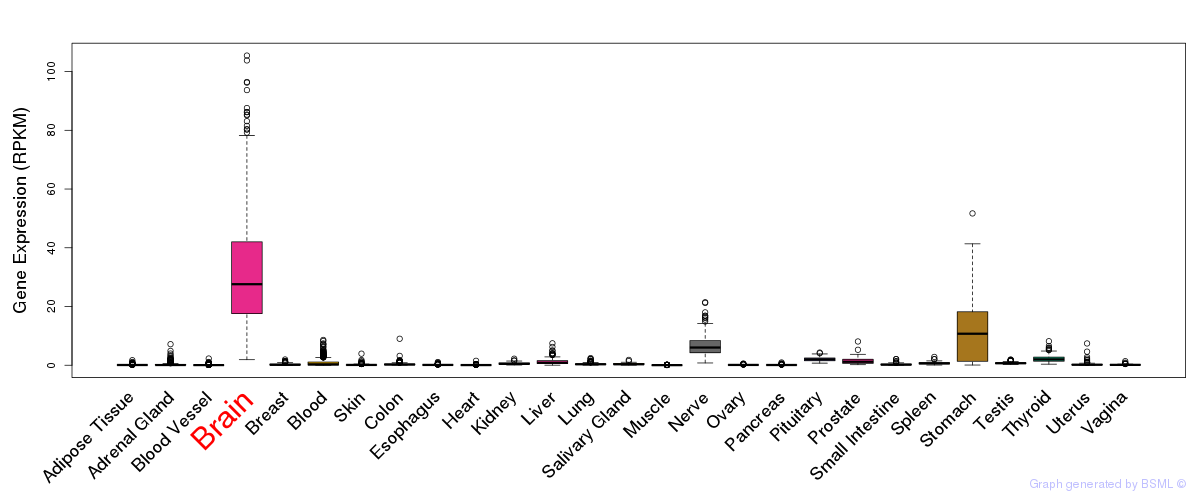
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SS18L1 | 0.96 | 0.95 |
| GPR173 | 0.95 | 0.94 |
| KDM4B | 0.95 | 0.94 |
| ZNF398 | 0.95 | 0.95 |
| PFKFB4 | 0.94 | 0.93 |
| ZNF74 | 0.94 | 0.93 |
| KDM2B | 0.94 | 0.95 |
| BCORL1 | 0.94 | 0.93 |
| BCL9 | 0.94 | 0.95 |
| SEMA4C | 0.94 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.72 | -0.78 |
| AF347015.31 | -0.71 | -0.89 |
| AF347015.27 | -0.70 | -0.88 |
| AIFM3 | -0.70 | -0.74 |
| MT-CO2 | -0.70 | -0.89 |
| S100B | -0.69 | -0.81 |
| HLA-F | -0.68 | -0.72 |
| AF347015.33 | -0.68 | -0.84 |
| FXYD1 | -0.68 | -0.84 |
| IFI27 | -0.67 | -0.84 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity | IEA | - | |
| GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005975 | carbohydrate metabolic process | TAS | 10783264 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | 10783264 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS CHONDROITIN SULFATE | 22 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS HEPARAN SULFATE | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| STARK BRAIN 22Q11 DELETION | 13 | 10 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-149 | 763 | 769 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-218 | 1379 | 1386 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.