Gene Page: GLI2
Summary ?
| GeneID | 2736 |
| Symbol | GLI2 |
| Synonyms | CJS|HPE9|PHS2|THP1|THP2 |
| Description | GLI family zinc finger 2 |
| Reference | MIM:165230|HGNC:HGNC:4318|Ensembl:ENSG00000074047|HPRD:01312|Vega:OTTHUMG00000153741 |
| Gene type | protein-coding |
| Map location | 2q14 |
| Pascal p-value | 0.314 |
| Fetal beta | 1.634 |
| DMG | 2 (# studies) |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.023 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLI2 | chr2 | 121554927 | G | A | NM_005270 | p.11E>K | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| ch.2.121457425R | 2 | 121740955 | GLI2 | 3.12E-5 | -0.685 | 0.019 | DMG:Wockner_2014 |
| cg21759953 | 2 | 121748257 | GLI2 | 5.31E-4 | 0.366 | 0.048 | DMG:Wockner_2014 |
| cg03575764 | 2 | 121493832 | GLI2 | 2.92E-8 | -0.01 | 9E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
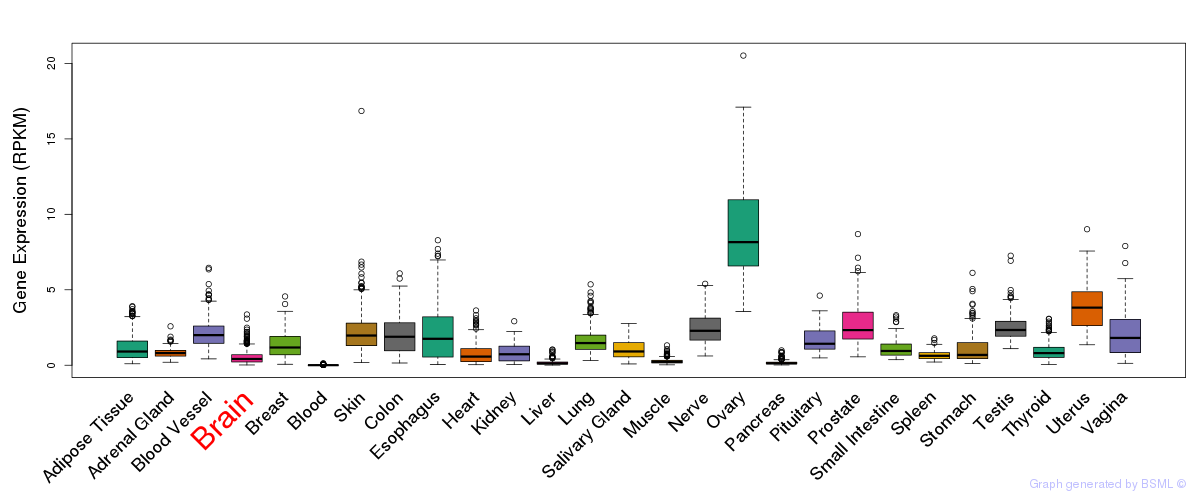
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 15994174 | |
| GO:0008270 | zinc ion binding | IDA | 8378770 | |
| GO:0016563 | transcription activator activity | IDA | 15175043 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IDA | 9557682 |15175043 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | ISS | axon (GO term level: 13) | - |
| GO:0048666 | neuron development | ISS | neuron (GO term level: 9) | - |
| GO:0021965 | spinal cord ventral commissure morphogenesis | ISS | neuron, axon (GO term level: 15) | - |
| GO:0030902 | hindbrain development | ISS | Brain (GO term level: 8) | - |
| GO:0021517 | ventral spinal cord development | ISS | neuron (GO term level: 8) | - |
| GO:0021508 | floor plate formation | ISS | neuron, Glial (GO term level: 10) | - |
| GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification | ISS | neuron (GO term level: 12) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IMP | 15994174 | |
| GO:0001501 | skeletal system development | ISS | - | |
| GO:0001822 | kidney development | ISS | - | |
| GO:0002076 | osteoblast development | ISS | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0009954 | proximal/distal pattern formation | ISS | - | |
| GO:0007224 | smoothened signaling pathway | IDA | 15994174 | |
| GO:0008283 | cell proliferation | IDA | 15994174 | |
| GO:0007507 | heart development | ISS | - | |
| GO:0007442 | hindgut morphogenesis | ISS | - | |
| GO:0007389 | pattern specification process | ISS | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | ISS | - | |
| GO:0021983 | pituitary gland development | ISS | - | |
| GO:0048754 | branching morphogenesis of a tube | ISS | - | |
| GO:0030879 | mammary gland development | ISS | - | |
| GO:0030324 | lung development | ISS | - | |
| GO:0035295 | tube development | ISS | - | |
| GO:0030903 | notochord development | ISS | - | |
| GO:0021513 | spinal cord dorsal/ventral patterning | ISS | - | |
| GO:0021696 | cerebellar cortex morphogenesis | ISS | - | |
| GO:0045740 | positive regulation of DNA replication | IDA | 12165851 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 12165851 |15994174 | |
| GO:0048566 | embryonic gut development | ISS | - | |
| GO:0048589 | developmental growth | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG 2PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU UP | 53 | 37 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 2 | 29 | 14 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-182 | 1410 | 1417 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-30-5p | 953 | 959 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.