Gene Page: GLRB
Summary ?
| GeneID | 2743 |
| Symbol | GLRB |
| Synonyms | HKPX2 |
| Description | glycine receptor beta |
| Reference | MIM:138492|HGNC:HGNC:4329|Ensembl:ENSG00000109738|HPRD:11751|Vega:OTTHUMG00000161954 |
| Gene type | protein-coding |
| Map location | 4q31.3 |
| Pascal p-value | 0.328 |
| Sherlock p-value | 0.959 |
| Fetal beta | -1.865 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10120856 | 4 | 157996959 | GLRB | 2.863E-4 | 0.382 | 0.039 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
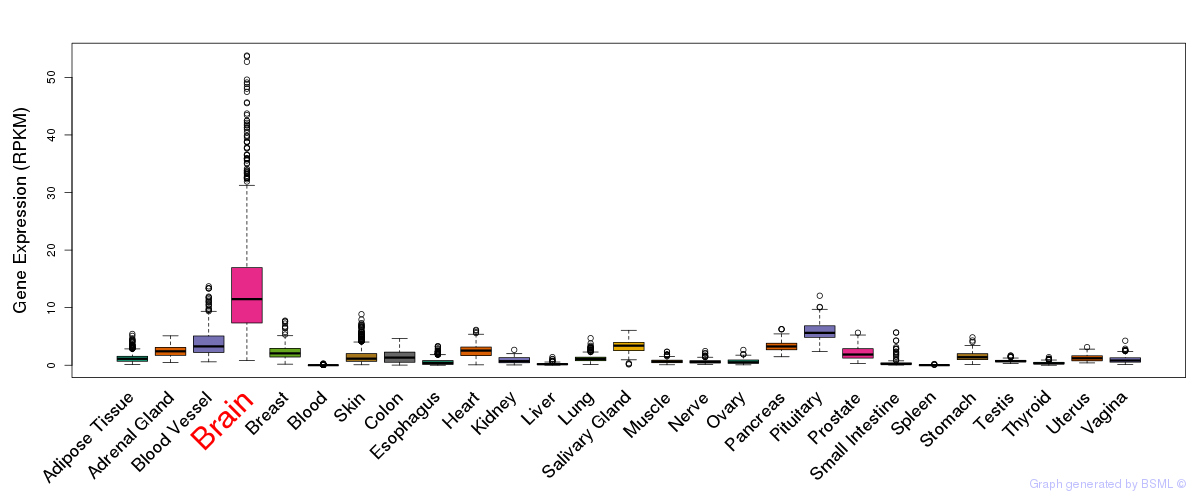
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TPP1 | 0.90 | 0.93 |
| C5orf4 | 0.88 | 0.87 |
| CDH20 | 0.87 | 0.86 |
| IGSF11 | 0.86 | 0.86 |
| GALNTL2 | 0.86 | 0.82 |
| GPRC5B | 0.85 | 0.83 |
| PADI2 | 0.85 | 0.84 |
| WIPF1 | 0.85 | 0.90 |
| KCNJ10 | 0.85 | 0.90 |
| ATP10B | 0.85 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.57 | -0.59 |
| AIM2 | -0.56 | -0.62 |
| KIAA1949 | -0.54 | -0.45 |
| STMN1 | -0.53 | -0.51 |
| MED19 | -0.53 | -0.56 |
| ZNF300 | -0.53 | -0.43 |
| TUBB2B | -0.53 | -0.50 |
| RBMX2 | -0.53 | -0.61 |
| CCDC28B | -0.53 | -0.60 |
| ZNF821 | -0.52 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0005515 | protein binding | IPI | 11929858 |15215304 | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0016934 | extracellular-glycine-gated chloride channel activity | IDA | 8717357 | |
| GO:0016594 | glycine binding | IMP | 15748848 | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | IMP | neuron, Synap, Neurotransmitter (GO term level: 6) | 11929858 |
| GO:0007218 | neuropeptide signaling pathway | IDA | Neurotransmitter (GO term level: 8) | 8717357 |
| GO:0007399 | nervous system development | IMP | neurite (GO term level: 5) | 11929858 |
| GO:0001964 | startle response | IMP | 11929858 | |
| GO:0006821 | chloride transport | IEA | - | |
| GO:0006811 | ion transport | IDA | 8717357 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | 8717357 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LIVER DN | 10 | 9 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 797 | 804 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-135 | 404 | 410 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-18 | 139 | 146 | 1A,m8 | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-203.1 | 33 | 39 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-369-3p | 435 | 441 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.