Gene Page: HINT1
Summary ?
| GeneID | 3094 |
| Symbol | HINT1 |
| Synonyms | HINT|NMAN|PKCI-1|PRKCNH1 |
| Description | histidine triad nucleotide binding protein 1 |
| Reference | MIM:601314|HGNC:HGNC:4912|Ensembl:ENSG00000169567|HPRD:03204|Vega:OTTHUMG00000128995 |
| Gene type | protein-coding |
| Map location | 5q31.2 |
| Pascal p-value | 0.664 |
| Sherlock p-value | 0.09 |
| Fetal beta | -0.288 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Expression | Meta-analysis of gene expression | P value: 2.914 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08523029 | 5 | 130500466 | HINT1 | 3.14E-5 | -0.363 | 0.019 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
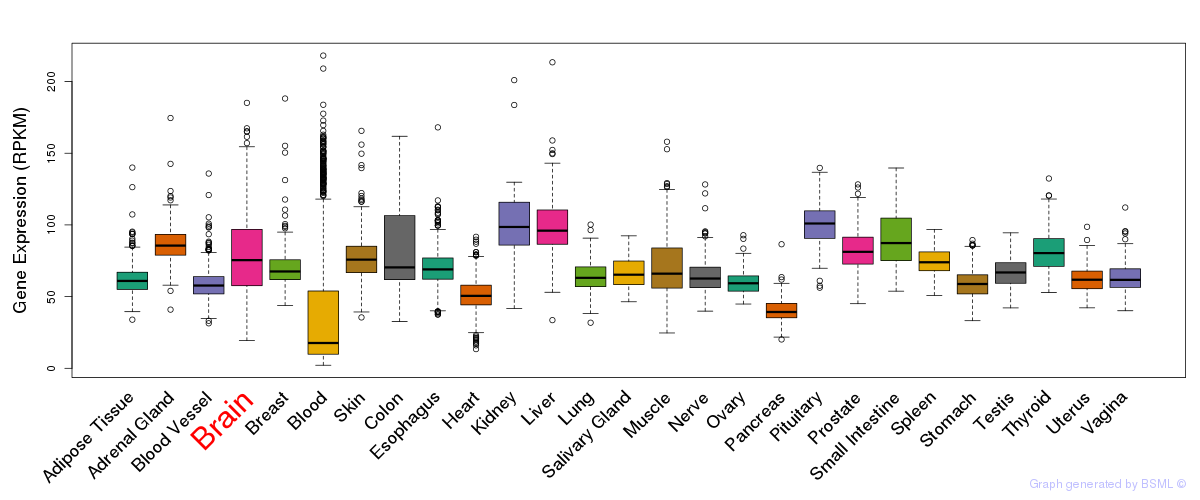
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARHGAP24 | 0.47 | 0.33 |
| CHRNA2 | 0.45 | 0.13 |
| MRVI1 | 0.45 | 0.37 |
| CHRNA6 | 0.45 | 0.22 |
| EPN3 | 0.44 | 0.18 |
| WIPI1 | 0.43 | 0.36 |
| RP4-539M6.2 | 0.42 | 0.29 |
| SUSD2 | 0.42 | 0.29 |
| EPS8L2 | 0.42 | 0.35 |
| CPT1A | 0.42 | 0.41 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARL4D | -0.29 | -0.28 |
| GPM6A | -0.28 | -0.27 |
| METTL9 | -0.28 | -0.32 |
| RNF5 | -0.27 | -0.32 |
| AC174470.1 | -0.27 | -0.35 |
| IER5L | -0.26 | -0.28 |
| VPS37D | -0.26 | -0.30 |
| ARPC5 | -0.26 | -0.25 |
| DRAP1 | -0.26 | -0.30 |
| STMN1 | -0.26 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005080 | protein kinase C binding | TAS | 9770345 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 8812426 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | TAS | 8812426 | |
| GO:0005634 | nucleus | TAS | 9770345 | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG DN | 45 | 29 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 3 | 28 | 16 | All SZGR 2.0 genes in this pathway |
| LUI TARGETS OF PAX8 PPARG FUSION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |