Gene Page: LYN
Summary ?
| GeneID | 4067 |
| Symbol | LYN |
| Synonyms | JTK8|p53Lyn|p56Lyn |
| Description | LYN proto-oncogene, Src family tyrosine kinase |
| Reference | MIM:165120|HGNC:HGNC:6735|HPRD:01301| |
| Gene type | protein-coding |
| Map location | 8q13 |
| Pascal p-value | 0.101 |
| Sherlock p-value | 0.514 |
| Fetal beta | -0.612 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0321 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16073549 | 8 | 56896784 | LYN | 2.43E-4 | 0.261 | 0.037 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10823579 | chr10 | 72348508 | LYN | 4067 | 0.1 | trans | ||
| rs10743139 | chr11 | 10425326 | LYN | 4067 | 0.19 | trans | ||
| rs11216720 | chr11 | 117909100 | LYN | 4067 | 0.07 | trans | ||
| rs12223165 | chr11 | 117910016 | LYN | 4067 | 0.07 | trans | ||
| rs11216730 | chr11 | 117918701 | LYN | 4067 | 0.03 | trans | ||
| rs16955618 | chr15 | 29937543 | LYN | 4067 | 1.074E-4 | trans |
Section II. Transcriptome annotation
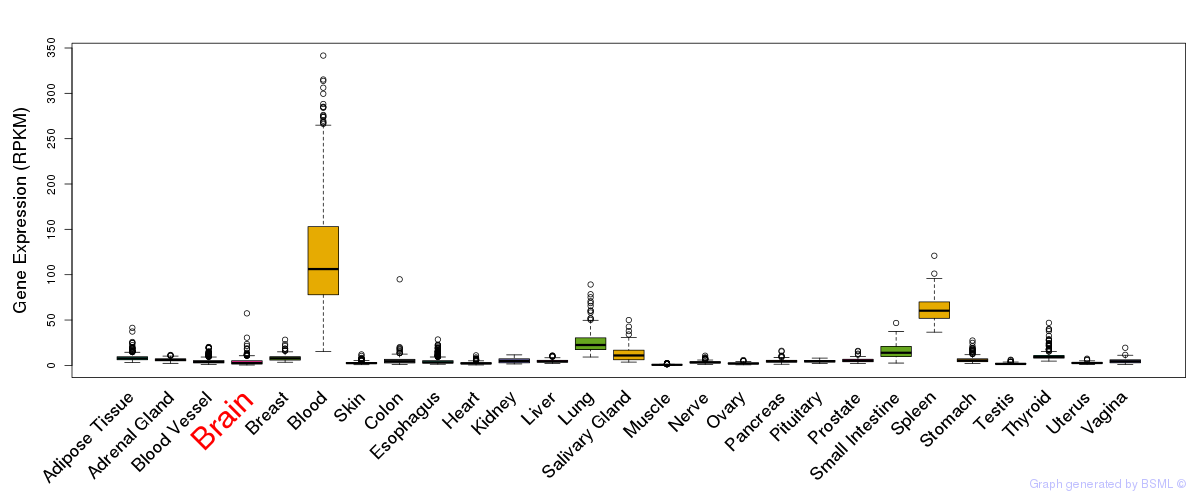
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRID1 | 0.83 | 0.84 |
| TAOK2 | 0.78 | 0.78 |
| SPTBN4 | 0.78 | 0.83 |
| KIAA0664 | 0.77 | 0.81 |
| ZBTB4 | 0.76 | 0.86 |
| C18orf1 | 0.76 | 0.81 |
| AC135592.2 | 0.76 | 0.78 |
| PCDH1 | 0.76 | 0.80 |
| RUNDC1 | 0.76 | 0.75 |
| AP006216.1 | 0.75 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf54 | -0.56 | -0.51 |
| AF347015.21 | -0.55 | -0.46 |
| CLEC2B | -0.52 | -0.47 |
| SYCP3 | -0.52 | -0.52 |
| GNG11 | -0.52 | -0.50 |
| C1orf61 | -0.52 | -0.49 |
| SAT1 | -0.51 | -0.49 |
| AF347015.31 | -0.50 | -0.43 |
| LGALS3 | -0.50 | -0.46 |
| VAMP5 | -0.50 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004716 | receptor signaling protein tyrosine kinase activity | TAS | 10748115 | |
| GO:0005515 | protein binding | IPI | 7859737 |8753773 |9020138 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | TAS | 3561390 | |
| GO:0007165 | signal transduction | TAS | 7650013 | |
| GO:0008284 | positive regulation of cell proliferation | ISS | - | |
| GO:0009725 | response to hormone stimulus | ISS | - | |
| GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | ISS | - | |
| GO:0030218 | erythrocyte differentiation | ISS | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 18029348 | |
| GO:0005634 | nucleus | ISS | - | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0005886 | plasma membrane | EXP | 10858437 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0045121 | membrane raft | IDA | 11313396 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BANK1 | BANK | FLJ20706 | FLJ34204 | B-cell scaffold protein with ankyrin repeats 1 | - | HPRD,BioGRID | 11782428 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 9581808 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 8058772 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Affinity Capture-Western | BioGRID | 12244174 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 12149655 |
| CBLC | CBL-3 | CBL-SL | RNF57 | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | - | HPRD,BioGRID | 10362357 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD | 7568175 |7589101 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | - | HPRD | 8647200 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | Affinity Capture-Western Biochemical Activity | BioGRID | 10228003 |10748054 |
| CD24 | CD24A | FLJ22950 | FLJ43543 | MGC75043 | CD24 molecule | - | HPRD,BioGRID | 8753773 |
| CD36 | CHDS7 | FAT | GP3B | GP4 | GPIV | PASIV | SCARB3 | CD36 molecule (thrombospondin receptor) | - | HPRD,BioGRID | 1715582 |
| CD72 | CD72b | LYB2 | CD72 molecule | Biochemical Activity | BioGRID | 9590210 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 8051175 |8910336 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD,BioGRID | 8806683 |10432313 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Affinity Capture-MS | BioGRID | 17353931 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 7636265 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD | 7636265|9973406 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | in vivo Protein-peptide | BioGRID | 7636265 |9973406 |
| CSF3R | CD114 | GCSFR | colony stimulating factor 3 receptor (granulocyte) | - | HPRD,BioGRID | 8197119 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | Two-hybrid | BioGRID | 14667819 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD,BioGRID | 9712716 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 9892651 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | Biochemical Activity Reconstituted Complex | BioGRID | 11071635 |11825908 |
| DOK3 | DOKL | FLJ22570 | FLJ39939 | docking protein 3 | - | HPRD | 10733577 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 9573010 |
| EVL | RNB6 | Enah/Vasp-like | - | HPRD,BioGRID | 10945997 |
| FCAR | CD89 | Fc fragment of IgA, receptor for | - | HPRD,BioGRID | 9427690 |
| FCER1G | FCRG | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | - | HPRD,BioGRID | 11388899 |
| FCGR2A | CD32 | CD32A | CDw32 | FCG2 | FCGR2 | FCGR2A1 | FcGR | IGFR2 | MGC23887 | MGC30032 | Fc fragment of IgG, low affinity IIa, receptor (CD32) | Affinity Capture-Western | BioGRID | 9268059 |
| FOLR1 | FBP | FOLR | FR-alpha | MOv18 | folate receptor 1 (adult) | - | HPRD | 10633085 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 11971018 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD,BioGRID | 11739737 |
| GALNAC4S-6ST | BRAG | DKFZp781H1369 | KIAA0598 | MGC34346 | RP11-47G11.1 | B cell RAG associated protein | - | HPRD | 10749872 |
| GP6 | GPIV | GPVI | MGC138168 | glycoprotein VI (platelet) | - | HPRD,BioGRID | 11943772 |
| GRIA3 | GLUR-C | GLUR-K3 | GLUR3 | GLURC | MRX94 | glutamate receptor, ionotrophic, AMPA 3 | - | HPRD | 9892356 |
| HCLS1 | CTTNL | HS1 | hematopoietic cell-specific Lyn substrate 1 | - | HPRD,BioGRID | 7682714 |
| IL7R | CD127 | CDW127 | IL-7R-alpha | IL7RA | ILRA | interleukin 7 receptor | - | HPRD | 7515933 |
| INPP5D | MGC104855 | MGC142140 | MGC142142 | SHIP | SHIP1 | SIP-145 | hp51CN | inositol polyphosphate-5-phosphatase, 145kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 12882960 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9573010 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | c-Kit interacts with p53Lyn. | BIND | 9341198 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 9341198 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | c-Kit interacts with p56Lyn. | BIND | 9341198 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD,BioGRID | 10026222 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | LYN autophosphorylates. This interaction was modeled on a demonstrated interaction using LYN from an unspecified species. | BIND | 12588871 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 11131153 |
| MME | CALLA | CD10 | DKFZp686O16152 | MGC126681 | MGC126707 | NEP | membrane metallo-endopeptidase | - | HPRD,BioGRID | 8943850 |
| MS4A2 | APY | ATOPY | FCER1B | FCERI | IGEL | IGER | IGHER | MS4A1 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | - | HPRD | 7528770 |7612892 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | Affinity Capture-Western Reconstituted Complex | BioGRID | 12750561 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD,BioGRID | 9020138 |
| NMT1 | NMT | N-myristoyltransferase 1 | - | HPRD,BioGRID | 11594778 |
| NPHS1 | CNF | NPHN | nephrosis 1, congenital, Finnish type (nephrin) | - | HPRD | 12846735 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 10790433 |
| PDE4A | DPDE2 | PDE4 | PDE46 | phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) | - | HPRD,BioGRID | 10206997 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | PDE4D4 interacts with Lyn SH3 domain. | BIND | 10571082 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 10858437 |
| PIK3CG | PI3CG | PI3K | PI3Kgamma | PIK3 | phosphoinositide-3-kinase, catalytic, gamma polypeptide | - | HPRD,BioGRID | 10318860 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD | 10586033 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8395016 |10981967 |
| PPP1R15A | GADD34 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | - | HPRD,BioGRID | 11517336 |
| PPP1R8 | ARD-1 | ARD1 | NIPP-1 | NIPP1 | PRO2047 | protein phosphatase 1, regulatory (inhibitor) subunit 8 | - | HPRD,BioGRID | 11104670 |
| PRAM1 | MGC39864 | PRAM-1 | PML-RARA regulated adaptor molecule 1 | - | HPRD,BioGRID | 11301322 |
| PRKCQ | MGC126514 | MGC141919 | PRKCT | nPKC-theta | protein kinase C, theta | - | HPRD | 11358993 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD,BioGRID | 9748231 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western | BioGRID | 9169439 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 11389892 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 11311138 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 10574931 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | Affinity Capture-Western Reconstituted Complex | BioGRID | 7516335 |
| RGS16 | A28-RGS14 | A28-RGS14P | RGS-R | regulator of G-protein signaling 16 | LYN phosphorylates RGS16. This interaction was modeled on a demonstrated interaction between LYN from an unspecified species and human RSG16. | BIND | 12588871 |
| RPL10 | DKFZp686J1851 | DXS648 | DXS648E | FLJ23544 | FLJ27072 | NOV | QM | ribosomal protein L10 | Reconstituted Complex | BioGRID | 12138090 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Biochemical Activity | BioGRID | 16640565 |
| RPS6KB2 | KLS | P70-beta | P70-beta-1 | P70-beta-2 | S6K-beta2 | S6K2 | SRK | STK14B | p70(S6K)-beta | p70S6Kb | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | Biochemical Activity | BioGRID | 16640565 |
| SH2B2 | APS | SH2B adaptor protein 2 | - | HPRD | 10872802 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 7650013 |
| SKAP1 | SCAP1 | SKAP55 | src kinase associated phosphoprotein 1 | - | HPRD,BioGRID | 9195899 |
| SKAP2 | MGC10411 | MGC33304 | PRAP | RA70 | SAPS | SCAP2 | SKAP-HOM | SKAP55R | src kinase associated phosphoprotein 2 | - | HPRD,BioGRID | 9837776 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Protein-peptide | BioGRID | 9169421 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 7831290 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | Affinity Capture-Western | BioGRID | 11071635 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | Reconstituted Complex | BioGRID | 10790433 |
| TRPV4 | OTRPC4 | TRP12 | VR-OAC | VRL-2 | VRL2 | VROAC | transient receptor potential cation channel, subfamily V, member 4 | - | HPRD,BioGRID | 12538589 |
| TYK2 | JTK1 | tyrosine kinase 2 | - | HPRD,BioGRID | 9633884 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | Lyn interacts with ubiquitin. | BIND | 11046148 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | - | HPRD,BioGRID | 12496276 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPHA4 PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID IL5 PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR1 PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF KIT SIGNALING | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PECAM1 INTERACTIONS | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CTLA4 INHIBITORY SIGNALING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF SIGNALING BY CBL | 18 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE TUMOR SAMPLE UP | 16 | 12 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP | 53 | 35 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS | 44 | 31 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCER KINOME GREEN | 16 | 14 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| TSUDA ALVEOLAR SOFT PART SARCOMA | 10 | 7 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-330 | 444 | 450 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-378 | 139 | 145 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.