Gene Page: MAB21L1
Summary ?
| GeneID | 4081 |
| Symbol | MAB21L1 |
| Synonyms | CAGR1|Nbla00126 |
| Description | mab-21-like 1 (C. elegans) |
| Reference | MIM:601280|HGNC:HGNC:6757|HPRD:03178| |
| Gene type | protein-coding |
| Map location | 13q13 |
| Fetal beta | 0.848 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | MAB21L1 | 4081 | 2.435E-21 | trans | ||
| rs3845734 | chr2 | 171125572 | MAB21L1 | 4081 | 0.07 | trans | ||
| rs7584986 | chr2 | 184111432 | MAB21L1 | 4081 | 8.99E-11 | trans | ||
| rs6741060 | chr2 | 217169088 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs9810143 | chr3 | 5060209 | MAB21L1 | 4081 | 0.12 | trans | ||
| rs2183142 | chr4 | 159232695 | MAB21L1 | 4081 | 6.661E-4 | trans | ||
| rs170776 | chr4 | 173276735 | MAB21L1 | 4081 | 0.01 | trans | ||
| rs1396222 | chr4 | 173279496 | MAB21L1 | 4081 | 0 | trans | ||
| rs335993 | chr4 | 173321789 | MAB21L1 | 4081 | 0 | trans | ||
| rs335980 | chr4 | 173329784 | MAB21L1 | 4081 | 0.01 | trans | ||
| rs335982 | chr4 | 173330945 | MAB21L1 | 4081 | 0.06 | trans | ||
| rs337984 | chr4 | 173411662 | MAB21L1 | 4081 | 0.04 | trans | ||
| rs10491487 | chr5 | 80323367 | MAB21L1 | 4081 | 5.758E-5 | trans | ||
| rs13191953 | chr6 | 24151042 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs12196880 | chr6 | 24313746 | MAB21L1 | 4081 | 0.17 | trans | ||
| rs10806988 | chr6 | 24341768 | MAB21L1 | 4081 | 0.11 | trans | ||
| rs9461864 | chr6 | 33481468 | MAB21L1 | 4081 | 0.03 | trans | ||
| rs16890367 | chr6 | 38078448 | MAB21L1 | 4081 | 0.03 | trans | ||
| rs1869293 | chr7 | 158181108 | MAB21L1 | 4081 | 0.18 | trans | ||
| rs9638062 | chr7 | 158221572 | MAB21L1 | 4081 | 0.18 | trans | ||
| rs6996695 | chr8 | 77540580 | MAB21L1 | 4081 | 0 | trans | ||
| rs3118341 | chr9 | 25185518 | MAB21L1 | 4081 | 0.06 | trans | ||
| rs1126130 | chr9 | 25209346 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs1998746 | chr9 | 25211761 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs1332433 | chr9 | 25214440 | MAB21L1 | 4081 | 0.2 | trans | ||
| rs9406868 | chr9 | 25223372 | MAB21L1 | 4081 | 9.325E-4 | trans | ||
| rs11139334 | chr9 | 84209393 | MAB21L1 | 4081 | 3.186E-4 | trans | ||
| rs4837782 | chr9 | 123330812 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs17143839 | chr10 | 8520836 | MAB21L1 | 4081 | 0.12 | trans | ||
| rs12098525 | chr10 | 87501764 | MAB21L1 | 4081 | 0.17 | trans | ||
| rs4122086 | chr11 | 92430845 | MAB21L1 | 4081 | 0.11 | trans | ||
| rs10501795 | chr11 | 92447417 | MAB21L1 | 4081 | 0.11 | trans | ||
| rs10501796 | chr11 | 92453155 | MAB21L1 | 4081 | 0.11 | trans | ||
| rs9989228 | chr14 | 37809252 | MAB21L1 | 4081 | 0.09 | trans | ||
| rs16955618 | chr15 | 29937543 | MAB21L1 | 4081 | 1.143E-23 | trans | ||
| rs2077735 | chr15 | 58479024 | MAB21L1 | 4081 | 0.04 | trans | ||
| rs9941296 | chr16 | 2759498 | MAB21L1 | 4081 | 0.01 | trans | ||
| rs16945437 | chr17 | 11797650 | MAB21L1 | 4081 | 0.04 | trans | ||
| rs11873184 | chr18 | 1584081 | MAB21L1 | 4081 | 0.13 | trans | ||
| rs11882889 | chr19 | 19887684 | MAB21L1 | 4081 | 0.11 | trans | ||
| rs1041786 | chr21 | 22617710 | MAB21L1 | 4081 | 0.01 | trans | ||
| rs16981387 | chr22 | 26495864 | MAB21L1 | 4081 | 0.15 | trans | ||
| rs16986332 | chrX | 10958354 | MAB21L1 | 4081 | 0.03 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
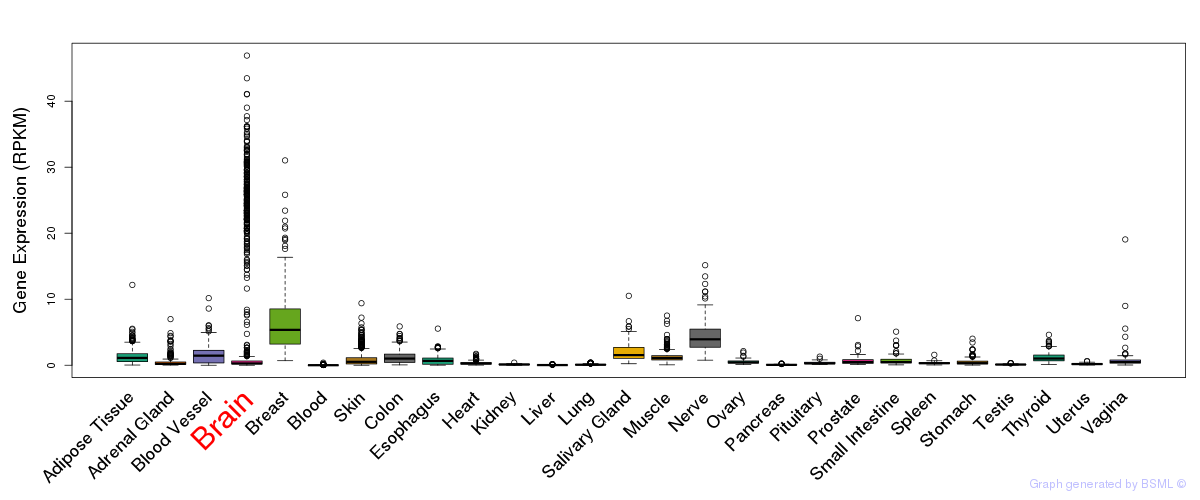
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 8733127 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 655 | 662 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-143 | 144 | 151 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-199 | 770 | 777 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-26 | 669 | 676 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-5p | 616 | 622 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-363 | 93 | 99 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-409-3p | 654 | 660 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-410 | 742 | 748 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| miR-452 | 94 | 101 | 1A,m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-543 | 676 | 682 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.