Gene Page: MYF6
Summary ?
| GeneID | 4618 |
| Symbol | MYF6 |
| Synonyms | CNM3|MRF4|bHLHc4|myf-6 |
| Description | myogenic factor 6 |
| Reference | MIM:159991|HGNC:HGNC:7566|Ensembl:ENSG00000111046|HPRD:01169|Vega:OTTHUMG00000170165 |
| Gene type | protein-coding |
| Map location | 12q21 |
| Pascal p-value | 0.919 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05860723 | 12 | 81102486 | MYF6 | 4.158E-4 | -0.285 | 0.044 | DMG:Wockner_2014 |
| cg05981335 | 12 | 81102562 | MYF6 | 5.723E-4 | -0.255 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
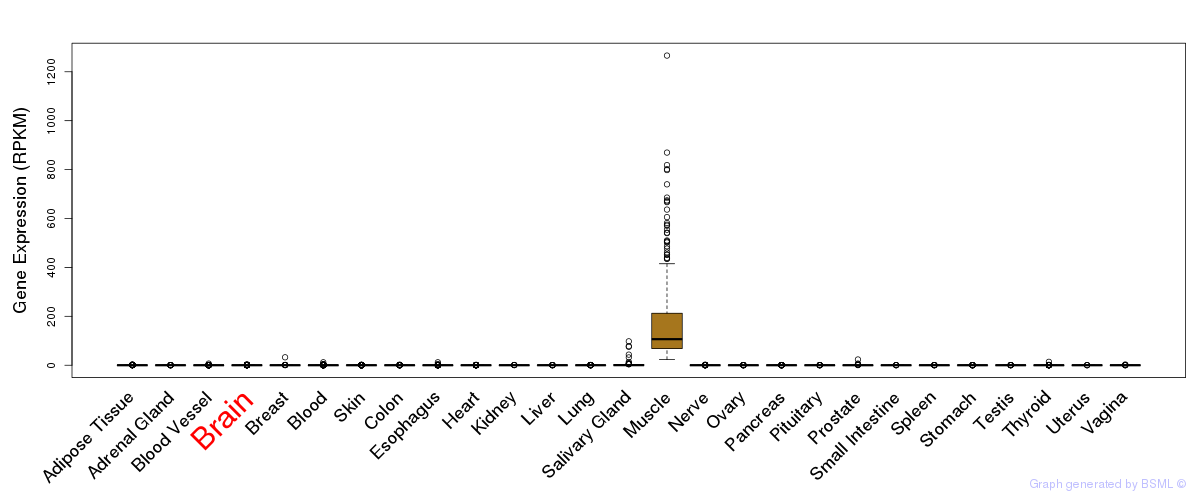
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDKL5 | 0.94 | 0.96 |
| FAM126B | 0.94 | 0.96 |
| PLEKHM3 | 0.94 | 0.95 |
| LONRF2 | 0.93 | 0.96 |
| HECW2 | 0.93 | 0.94 |
| C9orf126 | 0.93 | 0.92 |
| MYCBP2 | 0.92 | 0.92 |
| TMOD2 | 0.92 | 0.93 |
| LMTK2 | 0.92 | 0.93 |
| MAP2 | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.57 | -0.65 |
| DBI | -0.55 | -0.65 |
| RHOC | -0.55 | -0.65 |
| ACSF2 | -0.55 | -0.61 |
| EFEMP2 | -0.54 | -0.57 |
| EIF4EBP3 | -0.54 | -0.60 |
| C1orf61 | -0.54 | -0.74 |
| TLCD1 | -0.54 | -0.57 |
| FXYD1 | -0.53 | -0.57 |
| ENHO | -0.53 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 2311584 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | TAS | neuron (GO term level: 8) | 2311584 |
| GO:0001756 | somitogenesis | IEA | - | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 2311584 | |
| GO:0007517 | muscle development | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0016481 | negative regulation of transcription | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0045941 | positive regulation of transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 2311584 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FLI1 | 9 | 8 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH CHROMOSOME 12 TRISOMY | 24 | 12 | All SZGR 2.0 genes in this pathway |
| SUZUKI RESPONSE TO TSA AND DECITABINE 1B | 23 | 14 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| SUZUKI CTCFL TARGETS UP | 12 | 5 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-299-5p | 422 | 428 | m8 | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.