Gene Page: NDP
Summary ?
| GeneID | 4693 |
| Symbol | NDP |
| Synonyms | EVR2|FEVR|ND |
| Description | Norrie disease (pseudoglioma) |
| Reference | MIM:300658|HGNC:HGNC:7678|Ensembl:ENSG00000124479|HPRD:02404|Vega:OTTHUMG00000021391 |
| Gene type | protein-coding |
| Map location | Xp11.4 |
| Sherlock p-value | 0.511 |
| Fetal beta | 0.264 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
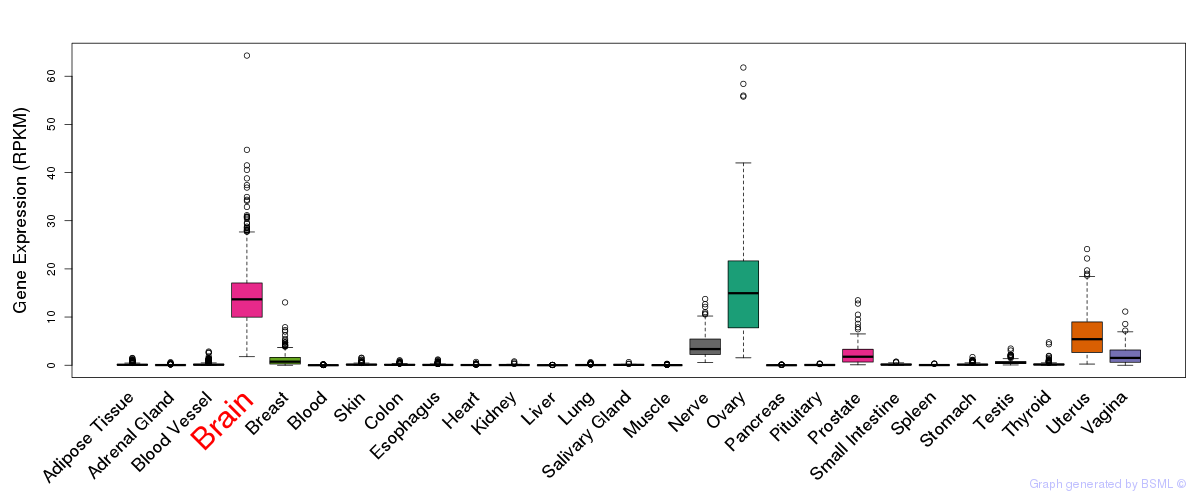
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDHX | 0.89 | 0.87 |
| EPS15 | 0.89 | 0.88 |
| NAPG | 0.88 | 0.89 |
| PRKAR1A | 0.88 | 0.85 |
| ARL8B | 0.87 | 0.88 |
| VAPB | 0.87 | 0.88 |
| SUCLA2 | 0.87 | 0.85 |
| ATP2A2 | 0.87 | 0.83 |
| USP32 | 0.87 | 0.85 |
| IDH3A | 0.87 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.50 | -0.30 |
| CLEC3B | -0.49 | -0.55 |
| IL32 | -0.48 | -0.36 |
| AL022328.1 | -0.46 | -0.46 |
| FAM159B | -0.46 | -0.46 |
| C1orf61 | -0.45 | -0.48 |
| AF347015.18 | -0.45 | -0.25 |
| C16orf74 | -0.44 | -0.41 |
| AC135724.1 | -0.43 | -0.44 |
| AC010300.1 | -0.43 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008083 | growth factor activity | TAS | 8298646 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 8252044 |
| GO:0001890 | placenta development | IEA | - | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 8298646 | |
| GO:0007165 | signal transduction | TAS | 8298646 | |
| GO:0007033 | vacuole organization | TAS | 10484772 | |
| GO:0008283 | cell proliferation | TAS | 8298646 | |
| GO:0007605 | sensory perception of sound | NAS | 8298646 | |
| GO:0007601 | visual perception | TAS | 10484772 | |
| GO:0050896 | response to stimulus | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 8298646 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC DN | 59 | 35 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| MATZUK IMPLANTATION AND UTERINE | 22 | 15 | All SZGR 2.0 genes in this pathway |
| PIONTEK PKD1 TARGETS DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |