Gene Page: NEUROD2
Summary ?
| GeneID | 4761 |
| Symbol | NEUROD2 |
| Synonyms | NDRF|bHLHa1 |
| Description | neuronal differentiation 2 |
| Reference | MIM:601725|HGNC:HGNC:7763|HPRD:03429| |
| Gene type | protein-coding |
| Map location | 17q12 |
| Pascal p-value | 3.437E-4 |
| Sherlock p-value | 0.043 |
| Fetal beta | 2.13 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01293647 | 17 | 37764264 | NEUROD2 | -0.027 | 0.31 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
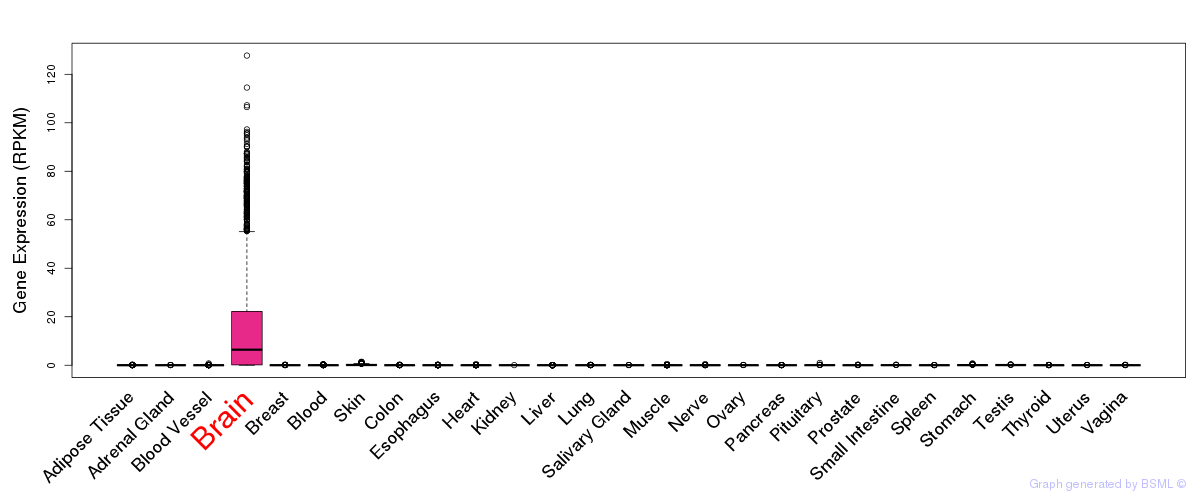
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HYAL2 | 0.68 | 0.71 |
| CDC42EP4 | 0.63 | 0.66 |
| SLC39A1 | 0.63 | 0.64 |
| MKNK2 | 0.63 | 0.60 |
| CORO1B | 0.62 | 0.63 |
| SEMA4B | 0.61 | 0.65 |
| XKR8 | 0.60 | 0.62 |
| MAFF | 0.60 | 0.62 |
| RBM38 | 0.60 | 0.60 |
| FADS2 | 0.60 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC104794.1 | -0.52 | -0.49 |
| RERGL | -0.51 | -0.50 |
| SYCP3 | -0.50 | -0.42 |
| USMG5 | -0.48 | -0.52 |
| C5orf53 | -0.47 | -0.46 |
| PIGB | -0.47 | -0.49 |
| SATL1 | -0.44 | -0.43 |
| UROS | -0.44 | -0.41 |
| MAEL | -0.44 | -0.46 |
| LY96 | -0.43 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 8816493 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 8816493 |
| GO:0001662 | behavioral fear response | IEA | - | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 8816493 | |
| GO:0008306 | associative learning | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 1509 | 1515 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-137 | 762 | 769 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-153 | 845 | 852 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 939 | 945 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-185 | 208 | 214 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-210 | 1171 | 1178 | 1A,m8 | hsa-miR-210 | CUGUGCGUGUGACAGCGGCUGA |
| miR-219 | 992 | 998 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-331 | 173 | 179 | m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-34/449 | 1027 | 1033 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-448 | 846 | 852 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-450 | 1509 | 1515 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-496 | 1660 | 1666 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.