Gene Page: NFKBIL1
Summary ?
| GeneID | 4795 |
| Symbol | NFKBIL1 |
| Synonyms | IKBL|LST1|NFKBIL |
| Description | NFKB inhibitor like 1 |
| Reference | MIM:601022|HGNC:HGNC:7800|Ensembl:ENSG00000204498|HPRD:07043|Vega:OTTHUMG00000031038 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 7.246E-6 |
| Sherlock p-value | 0.388 |
| Fetal beta | 0.073 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00585846 | 6 | 31515458 | NFKBIL1 | -0.021 | 0.66 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
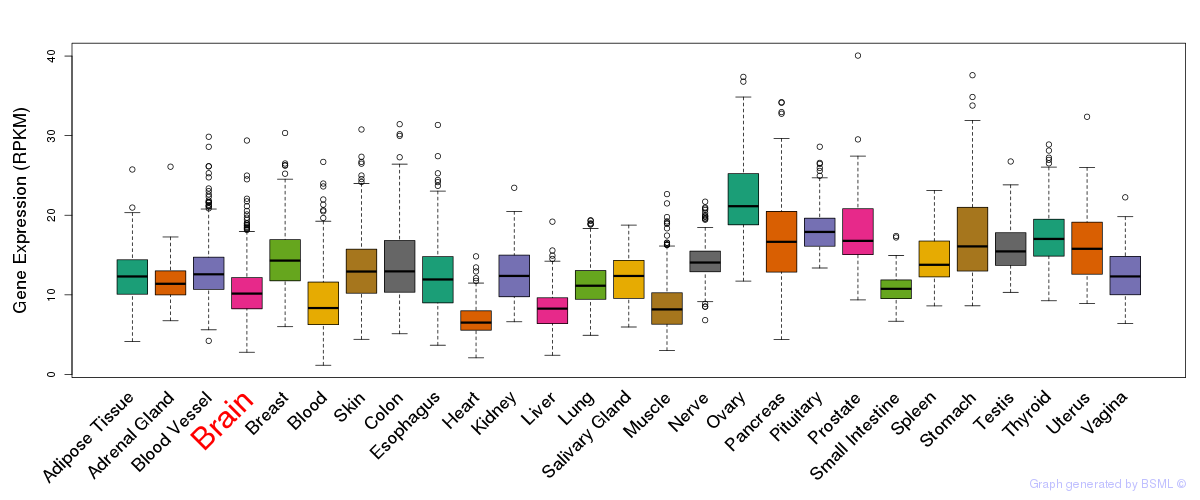
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR34 | 0.89 | 0.89 |
| GPC2 | 0.87 | 0.84 |
| MAD2L2 | 0.86 | 0.90 |
| GPX7 | 0.86 | 0.80 |
| DLL3 | 0.85 | 0.87 |
| FANCG | 0.85 | 0.88 |
| PYCR1 | 0.84 | 0.87 |
| RNASEH2A | 0.84 | 0.91 |
| PPP2R3B | 0.84 | 0.81 |
| CACNG4 | 0.84 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.60 | -0.74 |
| HLA-F | -0.58 | -0.74 |
| PTH1R | -0.58 | -0.71 |
| AF347015.27 | -0.57 | -0.86 |
| ALDOC | -0.57 | -0.70 |
| AIFM3 | -0.57 | -0.75 |
| AF347015.31 | -0.57 | -0.84 |
| AF347015.33 | -0.56 | -0.86 |
| TINAGL1 | -0.56 | -0.73 |
| MT-CO2 | -0.56 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042994 | cytoplasmic sequestering of transcription factor | TAS | 8081366 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IDA | 18029348 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST GAQ PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |