Gene Page: ROR1
Summary ?
| GeneID | 4919 |
| Symbol | ROR1 |
| Synonyms | NTRKR1|dJ537F10.1 |
| Description | receptor tyrosine kinase-like orphan receptor 1 |
| Reference | MIM:602336|HGNC:HGNC:10256|Ensembl:ENSG00000185483|HPRD:03821|Vega:OTTHUMG00000009022 |
| Gene type | protein-coding |
| Map location | 1p31.3 |
| Pascal p-value | 0.74 |
| Fetal beta | 0.517 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02692 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03768622 | 1 | 64239401 | ROR1 | 5.35E-5 | -0.627 | 0.022 | DMG:Wockner_2014 |
| cg04885825 | 1 | 64538523 | ROR1 | 7.86E-5 | -0.423 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs780587 | chr1 | 4505965 | ROR1 | 4919 | 0.02 | trans | ||
| rs2502827 | chr1 | 176044216 | ROR1 | 4919 | 0.15 | trans | ||
| rs16829545 | chr2 | 151977407 | ROR1 | 4919 | 7.474E-4 | trans | ||
| rs16841750 | chr2 | 158288461 | ROR1 | 4919 | 0.01 | trans | ||
| rs10183246 | chr2 | 237187882 | ROR1 | 4919 | 0.13 | trans | ||
| rs10195546 | chr2 | 237187933 | ROR1 | 4919 | 0.16 | trans | ||
| rs9810143 | chr3 | 5060209 | ROR1 | 4919 | 0.04 | trans | ||
| rs6797307 | chr3 | 8601563 | ROR1 | 4919 | 0.01 | trans | ||
| rs7692715 | chr4 | 125335209 | ROR1 | 4919 | 0.19 | trans | ||
| rs11937178 | chr4 | 148847598 | ROR1 | 4919 | 7.699E-4 | trans | ||
| rs11934409 | chr4 | 148848884 | ROR1 | 4919 | 7.699E-4 | trans | ||
| rs17136024 | chr7 | 54016166 | ROR1 | 4919 | 0.19 | trans | ||
| rs6467726 | chr7 | 137610394 | ROR1 | 4919 | 0.13 | trans | ||
| rs4921990 | chr8 | 18935646 | ROR1 | 4919 | 5.331E-4 | trans | ||
| rs10500981 | chr11 | 24543474 | ROR1 | 4919 | 0.09 | trans | ||
| rs504578 | chr11 | 78862291 | ROR1 | 4919 | 0.04 | trans | ||
| rs11564157 | chr12 | 40554096 | ROR1 | 4919 | 0.09 | trans | ||
| rs7488439 | chr12 | 105155814 | ROR1 | 4919 | 0.12 | trans | ||
| rs17036489 | chr12 | 105292283 | ROR1 | 4919 | 0.02 | trans | ||
| rs7337531 | chr13 | 109611085 | ROR1 | 4919 | 0 | trans | ||
| rs16955618 | chr15 | 29937543 | ROR1 | 4919 | 3.499E-6 | trans | ||
| rs4785436 | chr16 | 50590846 | ROR1 | 4919 | 0.19 | trans | ||
| rs8068272 | chr17 | 49952680 | ROR1 | 4919 | 0.01 | trans | ||
| rs17651763 | chr18 | 34232106 | ROR1 | 4919 | 0.01 | trans | ||
| rs17681878 | chr18 | 34767223 | ROR1 | 4919 | 2.137E-4 | trans | ||
| rs17782355 | chr19 | 55807585 | ROR1 | 4919 | 0.03 | trans |
Section II. Transcriptome annotation
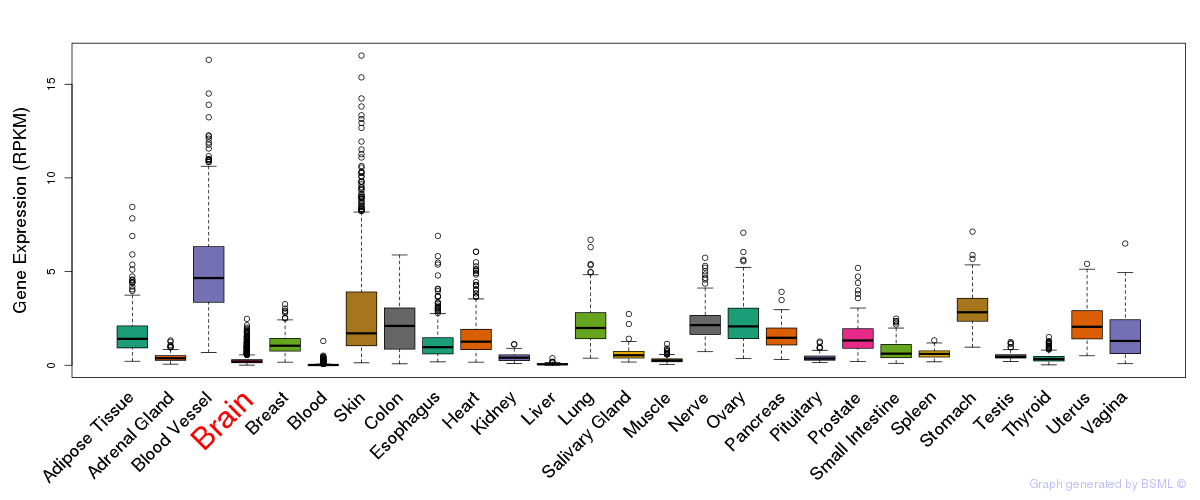
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MDS1 | 0.77 | 0.78 |
| EVI1 | 0.76 | 0.78 |
| COBLL1 | 0.75 | 0.78 |
| FZD6 | 0.75 | 0.72 |
| FLI1 | 0.75 | 0.76 |
| TEK | 0.74 | 0.78 |
| FLT1 | 0.73 | 0.75 |
| TGFBR2 | 0.72 | 0.80 |
| STAT3 | 0.72 | 0.74 |
| ZNF366 | 0.71 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNHG12 | -0.40 | -0.48 |
| ZNF32 | -0.38 | -0.42 |
| AC009171.1 | -0.37 | -0.41 |
| ACSM3 | -0.37 | -0.41 |
| PFDN5 | -0.37 | -0.45 |
| COMMD3 | -0.36 | -0.37 |
| RPL31 | -0.36 | -0.48 |
| RPL35 | -0.36 | -0.45 |
| NDUFAF2 | -0.36 | -0.38 |
| AC008763.1 | -0.36 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 8875995 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 1334494 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 8875995 | |
| GO:0005886 | plasma membrane | TAS | 1334494 |8875995 | |
| GO:0005887 | integral to plasma membrane | TAS | 1334494 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD DN | 36 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN | 50 | 32 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN | 56 | 39 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |