Gene Page: CLDN11
Summary ?
| GeneID | 5010 |
| Symbol | CLDN11 |
| Synonyms | OSP|OTM |
| Description | claudin 11 |
| Reference | MIM:601326|HGNC:HGNC:8514|HPRD:03207| |
| Gene type | protein-coding |
| Map location | 3q26.2-q26.3 |
| Pascal p-value | 0.017 |
| Sherlock p-value | 0.418 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs651185 | chr1 | 233871029 | CLDN11 | 5010 | 0.12 | trans |
Section II. Transcriptome annotation
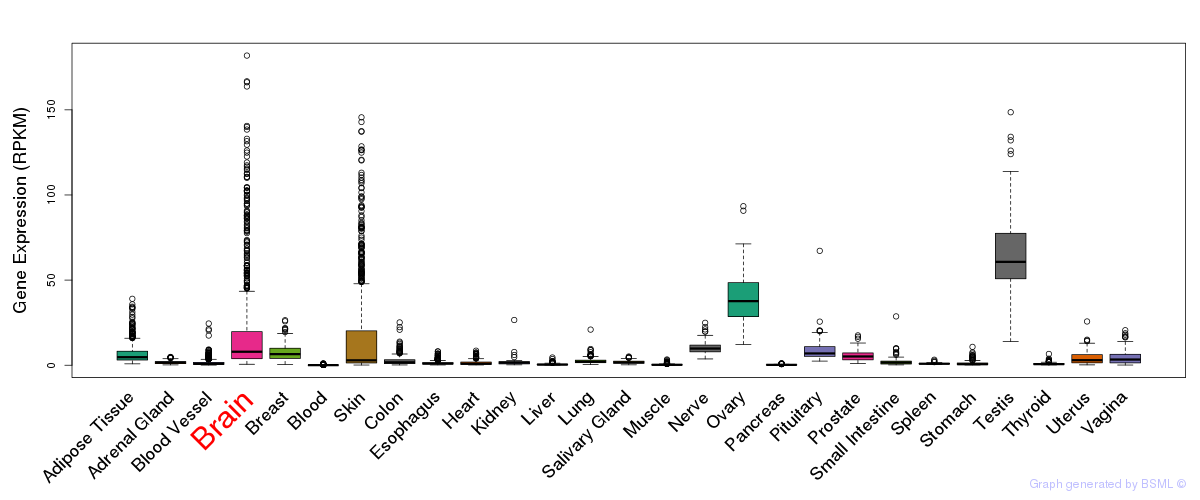
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GART | 0.88 | 0.85 |
| PABPCP5 | 0.88 | 0.89 |
| ZNF512 | 0.87 | 0.85 |
| TMEM57 | 0.87 | 0.85 |
| PRPSAP2 | 0.87 | 0.84 |
| DHX57 | 0.87 | 0.86 |
| XRN2 | 0.87 | 0.87 |
| TRAFD1 | 0.87 | 0.84 |
| CEP57 | 0.87 | 0.87 |
| ARFGAP3 | 0.87 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.71 | -0.77 |
| C5orf53 | -0.70 | -0.74 |
| AF347015.31 | -0.68 | -0.81 |
| MT-CO2 | -0.68 | -0.83 |
| AF347015.27 | -0.67 | -0.80 |
| AF347015.33 | -0.67 | -0.81 |
| AIFM3 | -0.67 | -0.69 |
| FBXO2 | -0.66 | -0.65 |
| CA4 | -0.66 | -0.72 |
| MT-CYB | -0.66 | -0.80 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005198 | structural molecule activity | IEA | - | |
| GO:0042802 | identical protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008366 | axon ensheathment | IEA | neuron, axon (GO term level: 12) | - |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007283 | spermatogenesis | IEA | - | |
| GO:0016338 | calcium-independent cell-cell adhesion | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | ISS | Brain (GO term level: 10) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME TIGHT JUNCTION INTERACTIONS | 29 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| ZIRN TRETINOIN RESPONSE WT1 UP | 23 | 14 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| PIONTEK PKD1 TARGETS DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WANG THOC1 TARGETS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 179 | 186 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 179 | 185 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-205 | 981 | 988 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-25/32/92/363/367 | 1314 | 1321 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.