Gene Page: OTX2
Summary ?
| GeneID | 5015 |
| Symbol | OTX2 |
| Synonyms | CPHD6|MCOPS5 |
| Description | orthodenticle homeobox 2 |
| Reference | MIM:600037|HGNC:HGNC:8522|HPRD:07190| |
| Gene type | protein-coding |
| Map location | 14q22.3 |
| Pascal p-value | 0.086 |
| Fetal beta | -0.131 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23316987 | 14 | 57272594 | OTX2 | 3.69E-6 | -0.484 | 0.009 | DMG:Wockner_2014 |
| cg24748769 | 14 | 57271015 | OTX2 | 1.636E-4 | -0.359 | 0.033 | DMG:Wockner_2014 |
| cg13411015 | 14 | 57274900 | OTX2 | 1.66E-8 | -0.023 | 6.1E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17091014 | chr14 | 56562713 | OTX2 | 5015 | 0.05 | cis | ||
| rs512189 | chr1 | 182632586 | OTX2 | 5015 | 0.12 | trans | ||
| rs6666913 | chr1 | 237092144 | OTX2 | 5015 | 0 | trans | ||
| rs16865199 | chr2 | 6621910 | OTX2 | 5015 | 0.13 | trans | ||
| rs16865228 | chr2 | 6629622 | OTX2 | 5015 | 0.1 | trans | ||
| rs16865230 | chr2 | 6630187 | OTX2 | 5015 | 0.1 | trans | ||
| rs11691188 | chr2 | 6631799 | OTX2 | 5015 | 0.1 | trans | ||
| rs13403353 | chr2 | 144067716 | OTX2 | 5015 | 0.18 | trans | ||
| rs7609917 | chr3 | 76094301 | OTX2 | 5015 | 0.01 | trans | ||
| rs2324466 | chr3 | 76100753 | OTX2 | 5015 | 8.178E-5 | trans | ||
| rs17013713 | chr3 | 76105150 | OTX2 | 5015 | 4.865E-5 | trans | ||
| rs17013722 | chr3 | 76107656 | OTX2 | 5015 | 8.178E-5 | trans | ||
| rs17013724 | chr3 | 76107684 | OTX2 | 5015 | 8.178E-5 | trans | ||
| rs179852 | chr3 | 172328734 | OTX2 | 5015 | 0.06 | trans | ||
| rs16831155 | chr3 | 173574763 | OTX2 | 5015 | 0.11 | trans | ||
| rs10015205 | chr4 | 26144278 | OTX2 | 5015 | 0.1 | trans | ||
| rs11734196 | chr4 | 102796071 | OTX2 | 5015 | 0.03 | trans | ||
| rs10212684 | chr4 | 102822506 | OTX2 | 5015 | 0.01 | trans | ||
| rs11937178 | chr4 | 148847598 | OTX2 | 5015 | 0.04 | trans | ||
| rs11934409 | chr4 | 148848884 | OTX2 | 5015 | 0.04 | trans | ||
| rs9986296 | chr5 | 3199318 | OTX2 | 5015 | 7.317E-4 | trans | ||
| rs17502163 | chr5 | 120026331 | OTX2 | 5015 | 0.09 | trans | ||
| rs2895773 | chr5 | 148114635 | OTX2 | 5015 | 0.14 | trans | ||
| rs10059267 | chr5 | 168676624 | OTX2 | 5015 | 0.13 | trans | ||
| rs10078402 | chr5 | 168678851 | OTX2 | 5015 | 0.13 | trans | ||
| rs7740062 | chr6 | 27705597 | OTX2 | 5015 | 0.13 | trans | ||
| rs200470 | chr6 | 27762186 | OTX2 | 5015 | 0.05 | trans | ||
| rs201005 | chr6 | 27804159 | OTX2 | 5015 | 0.1 | trans | ||
| rs200955 | chr6 | 27839311 | OTX2 | 5015 | 0.13 | trans | ||
| rs200959 | chr6 | 27842046 | OTX2 | 5015 | 0.13 | trans | ||
| rs839777 | chr6 | 27909656 | OTX2 | 5015 | 0.11 | trans | ||
| rs149898 | chr6 | 27958071 | OTX2 | 5015 | 0.11 | trans | ||
| rs156742 | chr6 | 27963191 | OTX2 | 5015 | 0.11 | trans | ||
| rs175955 | chr6 | 28059733 | OTX2 | 5015 | 0.11 | trans | ||
| rs1225593 | chr6 | 28150518 | OTX2 | 5015 | 0.11 | trans | ||
| rs195749 | chr6 | 37253745 | OTX2 | 5015 | 0.01 | trans | ||
| rs16896683 | chr6 | 43687846 | OTX2 | 5015 | 0.15 | trans | ||
| rs17073406 | chr6 | 144447390 | OTX2 | 5015 | 0.06 | trans | ||
| rs992435 | chr7 | 97050802 | OTX2 | 5015 | 0.11 | trans | ||
| rs1017329 | chr8 | 40281396 | OTX2 | 5015 | 0.13 | trans | ||
| rs16920368 | chr8 | 55392852 | OTX2 | 5015 | 0.01 | trans | ||
| rs16927318 | chr8 | 62365911 | OTX2 | 5015 | 0.07 | trans | ||
| rs348150 | chr8 | 73371619 | OTX2 | 5015 | 0.1 | trans | ||
| rs16938220 | chr8 | 73377940 | OTX2 | 5015 | 0.09 | trans | ||
| rs4734607 | chr8 | 102991441 | OTX2 | 5015 | 0.07 | trans | ||
| rs4545144 | chr8 | 139689307 | OTX2 | 5015 | 0.13 | trans | ||
| rs4742740 | chr9 | 101524552 | OTX2 | 5015 | 0.15 | trans | ||
| rs2069410 | chr12 | 56364644 | OTX2 | 5015 | 3.293E-6 | trans | ||
| rs11105124 | chr12 | 89375908 | OTX2 | 5015 | 0.16 | trans | ||
| rs9670212 | chr13 | 29070572 | OTX2 | 5015 | 0.05 | trans | ||
| rs17834315 | chr13 | 37991091 | OTX2 | 5015 | 5.564E-4 | trans | ||
| rs11841288 | chr13 | 60215833 | OTX2 | 5015 | 0.09 | trans | ||
| rs16946913 | chr15 | 63653889 | OTX2 | 5015 | 0.09 | trans | ||
| rs7174517 | chr15 | 87256320 | OTX2 | 5015 | 0.15 | trans | ||
| rs17694187 | chr15 | 91881259 | OTX2 | 5015 | 0.13 | trans | ||
| rs17142983 | chr16 | 7294508 | OTX2 | 5015 | 0 | trans | ||
| rs8056710 | chr16 | 7447629 | OTX2 | 5015 | 0.13 | trans | ||
| rs6499075 | chr16 | 66387395 | OTX2 | 5015 | 0.01 | trans | ||
| rs17736134 | chr18 | 70665805 | OTX2 | 5015 | 0.06 | trans | ||
| rs2469060 | chr18 | 70673106 | OTX2 | 5015 | 0.06 | trans | ||
| rs2469009 | chr18 | 70713334 | OTX2 | 5015 | 0.09 | trans | ||
| rs409642 | chr20 | 54250273 | OTX2 | 5015 | 0.02 | trans |
Section II. Transcriptome annotation
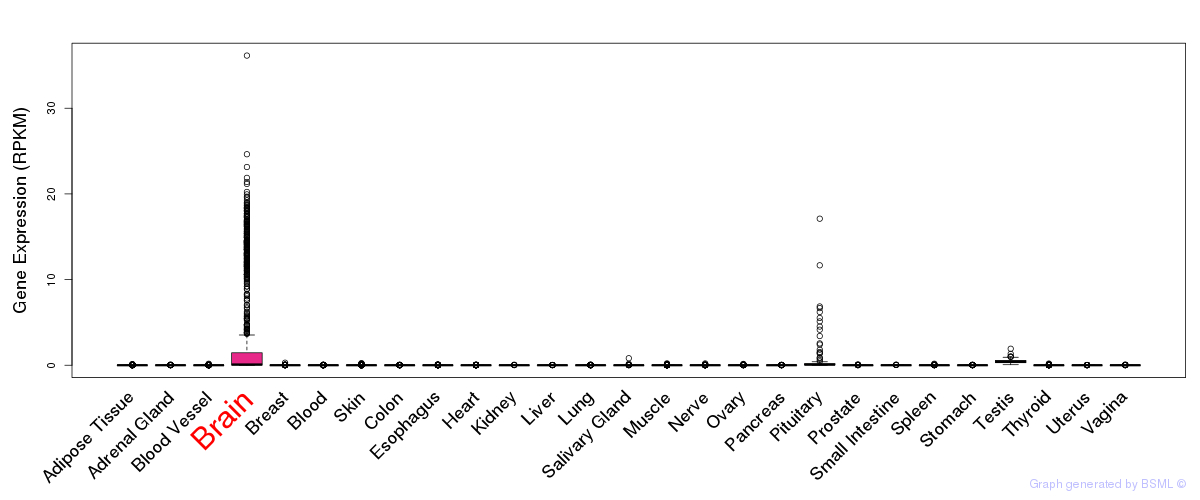
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | NAS | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008190 | eukaryotic initiation factor 4E binding | TAS | 15705863 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEA | Brain (GO term level: 6) | - |
| GO:0007411 | axon guidance | IDA | axon (GO term level: 13) | 16267555 |
| GO:0048663 | neuron fate commitment | IEA | neuron (GO term level: 9) | - |
| GO:0030901 | midbrain development | TAS | Brain (GO term level: 8) | 15705863 |
| GO:0030900 | forebrain development | TAS | Brain (GO term level: 8) | 15705863 |
| GO:0001708 | cell fate specification | IEA | - | |
| GO:0009953 | dorsal/ventral pattern formation | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0008589 | regulation of smoothened signaling pathway | TAS | 15705863 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0042472 | inner ear morphogenesis | IEA | - | |
| GO:0042706 | eye photoreceptor cell fate commitment | IEA | - | |
| GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | TAS | 15705863 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030426 | growth cone | IDA | axon, dendrite (GO term level: 5) | 16267555 |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| SASAI TARGETS OF CXCR6 AND PTCH1 UP | 13 | 9 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS UP | 112 | 65 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 51 | 58 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-128 | 621 | 627 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 503 | 509 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-138 | 171 | 177 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-142-5p | 509 | 515 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-148/152 | 450 | 456 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 641 | 647 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-219 | 558 | 565 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-27 | 621 | 627 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-3p | 452 | 459 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-374 | 697 | 703 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-410 | 955 | 962 | 1A,m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 641 | 647 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.