Gene Page: FURIN
Summary ?
| GeneID | 5045 |
| Symbol | FURIN |
| Synonyms | FUR|PACE|PCSK3|SPC1 |
| Description | furin, paired basic amino acid cleaving enzyme |
| Reference | MIM:136950|HGNC:HGNC:8568|Ensembl:ENSG00000140564|HPRD:00653|Vega:OTTHUMG00000149831 |
| Gene type | protein-coding |
| Map location | 15q26.1 |
| Pascal p-value | 3.671E-10 |
| Sherlock p-value | 0.749 |
| Fetal beta | -0.303 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1637 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs4702 | chr15 | 91426560 | AG | 2.298E-12 | UTR3 | FURIN | NM_002569:c.*1452A>G,NM_001289823:c.*1452A>G,NM_001289824:c.*1452A>G |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09169633 | 15 | 91447749 | FURIN | 9.823E-4 | -5.604 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
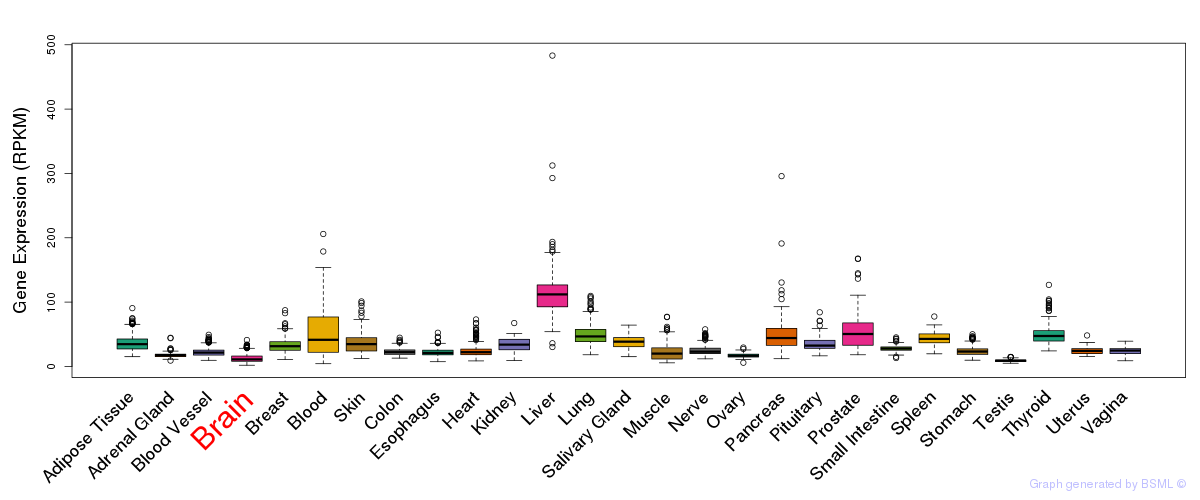
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0002020 | protease binding | IPI | 14744861 | |
| GO:0004867 | serine-type endopeptidase inhibitor activity | IDA | 10567353 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008233 | peptidase activity | IDA | 15082773 | |
| GO:0042277 | peptide binding | IDA | 8940009 | |
| GO:0048406 | nerve growth factor binding | IDA | 8615794 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006465 | signal peptide processing | IDA | 16912035 | |
| GO:0052548 | regulation of endopeptidase activity | IDA | 14744861 | |
| GO:0008283 | cell proliferation | IMP | 15899807 | |
| GO:0042176 | regulation of protein catabolic process | IMP | 15899807 | |
| GO:0051605 | protein maturation via proteolysis | IDA | 9242664 |10567353 |14744861 | |
| GO:0051605 | protein maturation via proteolysis | IMP | 15899807 | |
| GO:0019067 | viral assembly, maturation, egress, and release | IEP | 8940009 | |
| GO:0032804 | negative regulation of low-density lipoprotein receptor catabolic process | IDA | 16912035 | |
| GO:0032904 | negative regulation of nerve growth factor production | IDA | 10567353 | |
| GO:0032911 | negative regulation of transforming growth factor-beta1 production | IMP | 15899807 | |
| GO:0032940 | secretion by cell | IDA | 8615794 | |
| GO:0032455 | nerve growth factor processing | EXP | 12787574 | |
| GO:0043043 | peptide biosynthetic process | IDA | 8262946 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | EXP | 8615794 | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005802 | trans-Golgi network | IDA | 14744861 | |
| GO:0005615 | extracellular space | IDA | 10567353 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 16537537 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030140 | trans-Golgi network transport vesicle | IDA | 15078902 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NODAL | 18 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | 29 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRACELLULAR MATRIX ORGANIZATION | 87 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH PROCESSING IN GOLGI | 16 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE UP | 31 | 21 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 828 | 834 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124/506 | 1287 | 1293 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 1225 | 1231 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-137 | 365 | 372 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-15/16/195/424/497 | 829 | 835 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 1567 | 1574 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 42 | 48 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-185 | 431 | 437 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-219 | 1312 | 1319 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-22 | 249 | 255 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-24 | 724 | 730 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-331 | 1387 | 1393 | m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-448 | 1568 | 1574 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-452 | 1570 | 1576 | m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-543 | 1532 | 1538 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-9 | 56 | 62 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.