Gene Page: PCNT
Summary ?
| GeneID | 5116 |
| Symbol | PCNT |
| Synonyms | KEN|MOPD2|PCN|PCNT2|PCNTB|PCTN2|SCKL4 |
| Description | pericentrin |
| Reference | MIM:605925|HGNC:HGNC:16068|Ensembl:ENSG00000160299|HPRD:05804|Vega:OTTHUMG00000090665 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.708 |
| eGene | Frontal Cortex BA9 Myers' cis & trans Meta |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCNT | chr21 | 47831550 | C | G | NM_006031 | p.1855R>G | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7042553 | chr9 | 28380243 | PCNT | 5116 | 0.04 | trans | ||
| rs423974 | chr16 | 77930523 | PCNT | 5116 | 0.18 | trans |
Section II. Transcriptome annotation
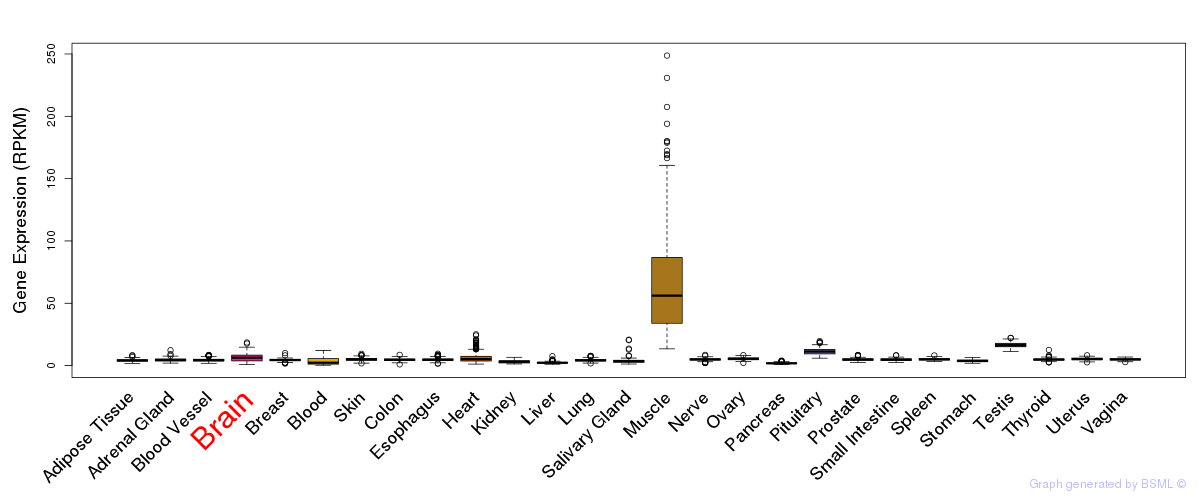
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CD300A | 0.52 | 0.42 |
| C2 | 0.49 | 0.44 |
| COL4A2 | 0.49 | 0.45 |
| STRA6 | 0.48 | 0.46 |
| HAPLN3 | 0.47 | 0.42 |
| MEST | 0.47 | 0.48 |
| MMP2 | 0.47 | 0.47 |
| VSIG4 | 0.47 | 0.36 |
| COL4A1 | 0.47 | 0.45 |
| PLVAP | 0.47 | 0.35 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.35 | -0.41 |
| ASPA | -0.34 | -0.39 |
| PLEKHB1 | -0.34 | -0.36 |
| CSRP1 | -0.34 | -0.35 |
| FBXO2 | -0.34 | -0.35 |
| AF347015.33 | -0.34 | -0.40 |
| HLA-F | -0.34 | -0.33 |
| ALDH1A1 | -0.34 | -0.35 |
| CA2 | -0.33 | -0.34 |
| HHATL | -0.33 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005516 | calmodulin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007051 | spindle organization | IEA | - | |
| GO:0042384 | cilium biogenesis | IDA | 15337773 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005814 | centriole | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 21Q22 AMPLICON | 16 | 11 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |