| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION
| 267 | 161 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP
| 290 | 177 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN
| 186 | 114 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP
| 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP
| 238 | 144 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP
| 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP
| 58 | 40 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP
| 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP
| 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN
| 75 | 50 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP
| 55 | 40 | All SZGR 2.0 genes in this pathway |
| CREIGHTON AKT1 SIGNALING VIA MTOR DN
| 23 | 16 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP
| 121 | 72 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER PROGRESSION UP
| 9 | 6 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP
| 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN
| 79 | 54 | All SZGR 2.0 genes in this pathway |
| MCBRYAN TERMINAL END BUD UP
| 12 | 12 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK
| 74 | 44 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP
| 88 | 58 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP
| 83 | 51 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM3
| 70 | 37 | All SZGR 2.0 genes in this pathway |
| FUJIWARA PARK2 IN LIVER CANCER UP
| 8 | 7 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA
| 164 | 118 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 MCF10A
| 20 | 13 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16P13 AMPLICON
| 120 | 49 | All SZGR 2.0 genes in this pathway |
| ZEILSTRA CD44 TARGETS DN
| 7 | 5 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP
| 126 | 84 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS
| 212 | 121 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS
| 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC
| 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC
| 58 | 43 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| JEPSEN SMRT TARGETS
| 33 | 23 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G
| 95 | 62 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G
| 171 | 96 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP
| 180 | 114 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP
| 115 | 80 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR REJECTION DN
| 10 | 6 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP
| 871 | 505 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN
| 138 | 92 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN
| 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP
| 212 | 128 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE
| 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE
| 81 | 66 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN
| 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2
| 277 | 172 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP
| 93 | 62 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN
| 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN
| 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN
| 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN
| 40 | 29 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP
| 127 | 78 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP
| 387 | 225 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B
| 549 | 316 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN
| 315 | 215 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP
| 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP
| 135 | 96 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP
| 199 | 143 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF
| 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS UP
| 48 | 33 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
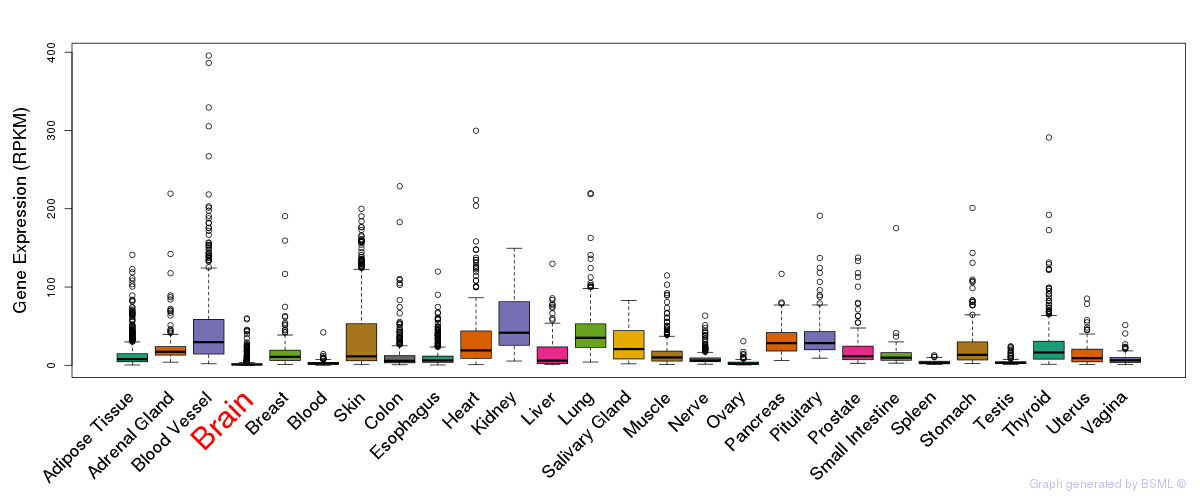
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation