Gene Page: AMOTL2
Summary ?
| GeneID | 51421 |
| Symbol | AMOTL2 |
| Synonyms | LCCP |
| Description | angiomotin like 2 |
| Reference | MIM:614658|HGNC:HGNC:17812|Ensembl:ENSG00000114019|HPRD:16485|Vega:OTTHUMG00000159777 |
| Gene type | protein-coding |
| Map location | 3q21-q22 |
| Pascal p-value | 0.298 |
| Fetal beta | 0.214 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06503171 | 3 | 134083286 | AMOTL2 | 1.258E-4 | -0.364 | 0.03 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
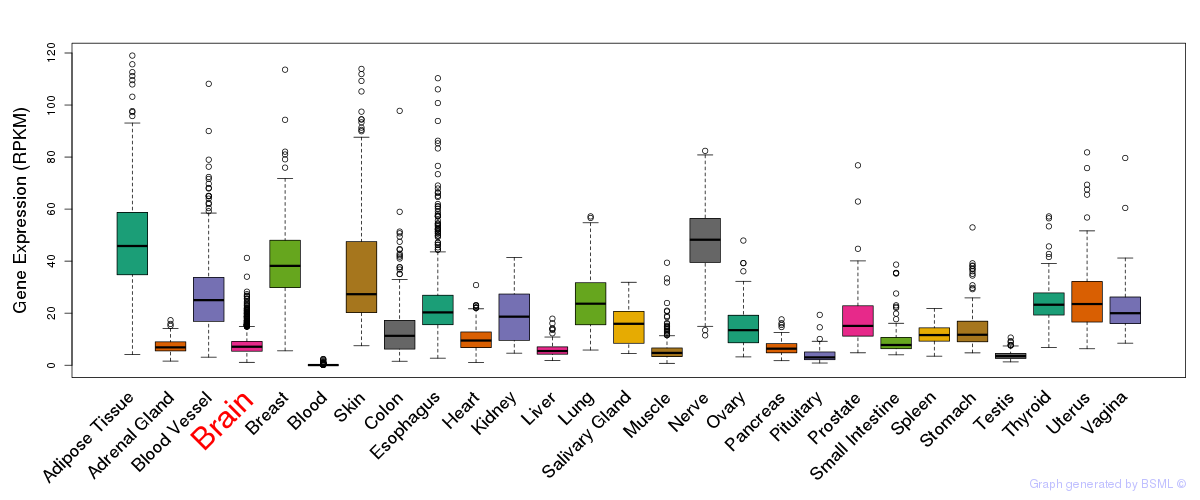
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042802 | identical protein binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY HIPPO | 22 | 15 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY UP | 33 | 18 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE UP | 34 | 27 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 1582 | 1589 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-145 | 1033 | 1039 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-149 | 942 | 949 | 1A,m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-150 | 1083 | 1090 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-182 | 1678 | 1684 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 1798 | 1804 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-224 | 1276 | 1282 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-24 | 955 | 961 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG | ||||
| miR-27 | 1664 | 1670 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 1546 | 1553 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-324-3p | 1273 | 1279 | 1A | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-381 | 638 | 644 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-382 | 1608 | 1615 | 1A,m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-9 | 1680 | 1686 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 1678 | 1684 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.