Gene Page: PML
Summary ?
| GeneID | 5371 |
| Symbol | PML |
| Synonyms | MYL|PP8675|RNF71|TRIM19 |
| Description | promyelocytic leukemia |
| Reference | MIM:102578|HGNC:HGNC:9113|Ensembl:ENSG00000140464|HPRD:00023|Vega:OTTHUMG00000137607 |
| Gene type | protein-coding |
| Map location | 15q22 |
| Pascal p-value | 0.329 |
| TADA p-value | 0.019 |
| Fetal beta | -0.311 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0455 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PML | chr15 | 74290439 | C | T | NM_002675 NM_033238 NM_033239 NM_033240 NM_033244 NM_033246 NM_033247 NM_033249 NM_033250 | p.75T>M p.75T>M p.75T>M p.75T>M p.75T>M p.75T>M p.75T>M p.75T>M p.75T>M | missense missense missense missense missense missense missense missense missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
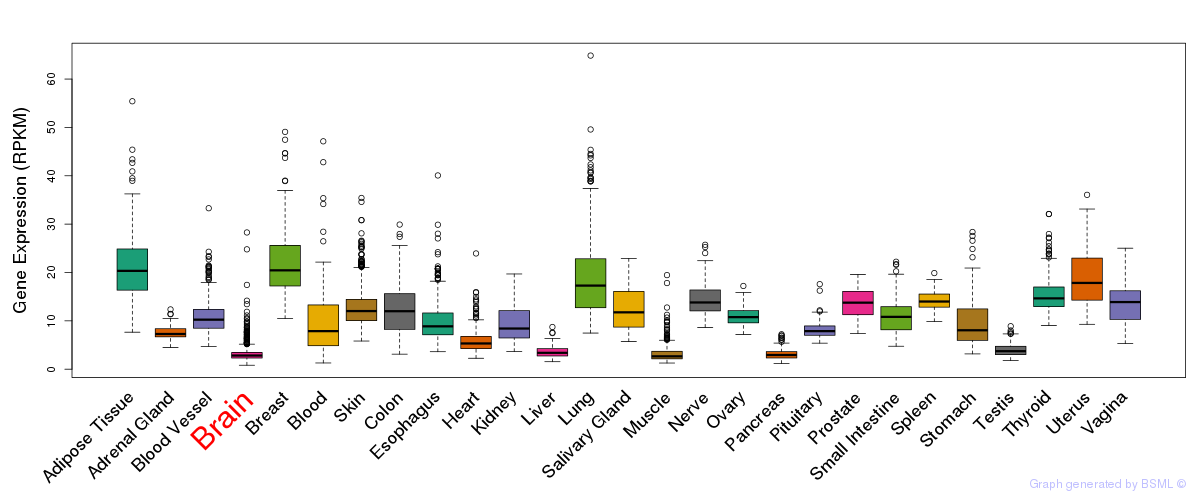
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | NAS | 1652369 | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9294197 |9671405 |11259576 |11432836 |12915590 |14976184 |15626733 |16915281 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008270 | zinc ion binding | NAS | 1652369 | |
| GO:0016564 | transcription repressor activity | IDA | 11432836 | |
| GO:0032183 | SUMO binding | IPI | 17081985 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001666 | response to hypoxia | IDA | 16915281 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006461 | protein complex assembly | IDA | 12915590 | |
| GO:0006281 | DNA repair | NAS | 12773567 | |
| GO:0006917 | induction of apoptosis | IMP | 16912307 | |
| GO:0010212 | response to ionizing radiation | IDA | 12773567 | |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | ISS | - | |
| GO:0016481 | negative regulation of transcription | IDA | 9448006 | |
| GO:0016525 | negative regulation of angiogenesis | IMP | 16915281 | |
| GO:0042771 | DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | ISS | - | |
| GO:0030308 | negative regulation of cell growth | IDA | 9448006 | |
| GO:0030578 | PML body organization | IMP | 17081985 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0051457 | maintenance of protein location in nucleus | IDA | 17332504 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005626 | insoluble fraction | IDA | 12915590 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IDA | 12915590 | |
| GO:0005730 | nucleolus | IDA | 15195100 | |
| GO:0005737 | cytoplasm | IDA | 16778193 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016605 | PML body | IDA | 9448006 |17081985 | |
| GO:0016605 | PML body | IDA | 10910364 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANKRD2 | ARPP | MGC104314 | ankyrin repeat domain 2 (stretch responsive muscle) | - | HPRD,BioGRID | 15136035 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | - | HPRD,BioGRID | 12354770 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | Reconstituted Complex | BioGRID | 9671405 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | ATR phosphorylates PML. | BIND | 15195100 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | PML3 interacts with Aurora A. | BIND | 15749021 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | Two-hybrid | BioGRID | 11175338 |
| CCNT1 | CCNT | CYCT1 | cyclin T1 | - | HPRD,BioGRID | 12727882 |
| CHFR | FLJ10796 | FLJ33629 | RNF116 | RNF196 | checkpoint with forkhead and ring finger domains | CHFR interacts an unspecified isoform of PML. | BIND | 15467728 |
| CHFR | FLJ10796 | FLJ33629 | RNF116 | RNF196 | checkpoint with forkhead and ring finger domains | - | HPRD,BioGRID | 15467728 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD | 10077561 |10610177 |12142048 |12234245 |12622724 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 10077561 |10610177 |12622724 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD | 10525530 |10669754 |10684855 |12595526 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Affinity Capture-Western Co-localization Two-hybrid | BioGRID | 10525530 |10669754 |10684855 |11244500 |
| EIF4E | CBP | EIF4E1 | EIF4EL1 | EIF4F | MGC111573 | eukaryotic translation initiation factor 4E | - | HPRD,BioGRID | 11500381 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD,BioGRID | 9671405 |
| GATA2 | MGC2306 | NFE1B | GATA binding protein 2 | - | HPRD,BioGRID | 10938104 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 11259576 |11430826 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 11259576 |11430826 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11259576 |
| HHEX | HEX | HMPH | HOX11L-PEN | PRH | PRHX | hematopoietically expressed homeobox | - | HPRD,BioGRID | 10597310 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 9671405 |
| MAPK11 | P38B | P38BETA2 | PRKM11 | SAPK2 | SAPK2B | p38-2 | p38Beta | mitogen-activated protein kinase 11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15273249 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD | 15273249 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | PML interacts with Mdm2. | BIND | 15195100 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 12759344 |12915590 |14507915 |15195100 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | - | HPRD | 11430826 |
| MYB | Cmyb | c-myb | c-myb_CDS | efg | v-myb myeloblastosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 15194430 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Myc interacts with PML. This interaction was modelled on a demonstrated interaction between mouse Myc and human PML. | BIND | 15735755 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 11430826 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 11585900 |
| NR4A1 | GFRP1 | HMR | MGC9485 | N10 | NAK-1 | NGFIB | NP10 | NUR77 | TR3 | nuclear receptor subfamily 4, group A, member 1 | - | HPRD,BioGRID | 12032831 |
| PAWR | PAR4 | Par-4 | PRKC, apoptosis, WT1, regulator | - | HPRD | 12717420 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Far Western | BioGRID | 10669754 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | - | HPRD | 12935927 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 10610177 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 9448006 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 12540841 |
| RPL11 | GIG34 | ribosomal protein L11 | PML interacts with L11 in the nucleolus. | BIND | 15195100 |
| RPL11 | GIG34 | ribosomal protein L11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15195100 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex | BioGRID | 10610177 |
| SENP2 | AXAM2 | DKFZp762A2316 | KIAA1331 | SMT3IP2 | SUMO1/sentrin/SMT3 specific peptidase 2 | - | HPRD | 12419228 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11430826 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | - | HPRD,BioGRID | 11430826 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 15356634 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | cPML interacts with Smad2. | BIND | 15356634 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | cPML interacts with Smad3. | BIND | 15356634 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 15356634 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 9819401 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD,BioGRID | 12622724 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12506013 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD | 8806687 |11080164 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Affinity Capture-Western in vitro in vivo Two-hybrid | BioGRID | 8806687 |9452416 |11948183 |
| SUMO2 | HSMT3 | MGC117191 | SMT3B | SMT3H2 | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | - | HPRD | 9452416 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 15569683 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD | 15356634 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | cPML interacts with T-beta-RI. | BIND | 15356634 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | cPML interacts with T-beta-RII. | BIND | 15356634 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD | 15356634 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | - | HPRD | 10610177 |
| TOPBP1 | TOP2BP1 | topoisomerase (DNA) II binding protein 1 | - | HPRD,BioGRID | 12773567 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11025664 |11080164 |12915590 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 11025664 |11080164 |11704853 |
| TP73 | P73 | tumor protein p73 | - | HPRD | 11832207 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD,BioGRID | 10610177 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | - | HPRD,BioGRID | 9570750 |
| TRIM69 | HSD34 | RNF36 | Trif | tripartite motif-containing 69 | - | HPRD,BioGRID | 12837286 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Biochemical Activity | BioGRID | 18691969 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 9294197 |11175338 |
| ZFYVE9 | MADHIP | NSP | SARA | SMADIP | zinc finger, FYVE domain containing 9 | - | HPRD,BioGRID | 15356634 |
| ZFYVE9 | MADHIP | NSP | SARA | SMADIP | zinc finger, FYVE domain containing 9 | cPML interacts with SARA. | BIND | 15356634 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PML PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS YELLOW DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| VETTER TARGETS OF PRKCA AND ETS1 UP | 16 | 8 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| OXFORD RALB TARGETS DN | 9 | 6 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP | 53 | 35 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR UP | 47 | 39 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C5 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |