Gene Page: PPT1
Summary ?
| GeneID | 5538 |
| Symbol | PPT1 |
| Synonyms | CLN1|INCL|PPT |
| Description | palmitoyl-protein thioesterase 1 |
| Reference | MIM:600722|HGNC:HGNC:9325|Ensembl:ENSG00000131238|HPRD:07203|Vega:OTTHUMG00000004495 |
| Gene type | protein-coding |
| Map location | 1p32 |
| Pascal p-value | 0.835 |
| Sherlock p-value | 0.316 |
| Fetal beta | -1.213 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 11 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19736671 | 1 | 40562678 | PPT1 | 4.55E-8 | -0.007 | 1.24E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
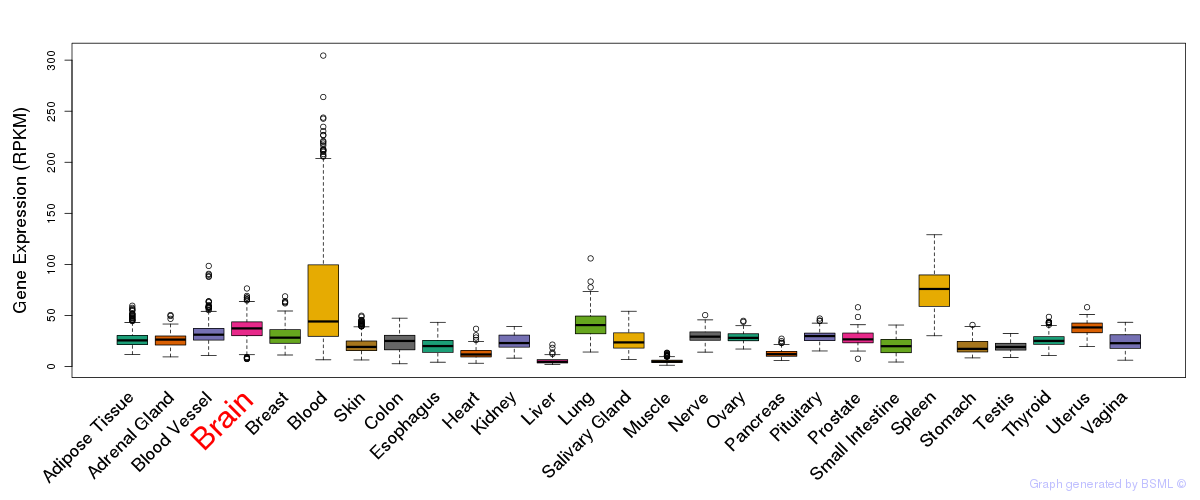
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NECAB2 | 0.80 | 0.78 |
| CNTN6 | 0.79 | 0.65 |
| ANXA3 | 0.78 | 0.53 |
| CALB2 | 0.77 | 0.55 |
| EGFL6 | 0.75 | 0.53 |
| NEFL | 0.73 | 0.68 |
| DOK7 | 0.71 | 0.59 |
| OTX2 | 0.71 | -0.04 |
| TP53I3 | 0.70 | 0.08 |
| SLC17A6 | 0.69 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FOXG1B | -0.27 | -0.36 |
| SMTN | -0.26 | -0.53 |
| ISLR2 | -0.25 | -0.27 |
| SIGIRR | -0.25 | -0.43 |
| ZEB2 | -0.25 | -0.32 |
| LZTS1 | -0.24 | -0.27 |
| TTC28 | -0.24 | -0.34 |
| EMX1 | -0.24 | -0.22 |
| MPPED1 | -0.24 | -0.24 |
| NEUROD2 | -0.24 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008474 | palmitoyl-(protein) hydrolase activity | IDA | 10658183 |10737604 | |
| GO:0008474 | palmitoyl-(protein) hydrolase activity | IEA | - | |
| GO:0008474 | palmitoyl-(protein) hydrolase activity | IMP | 7637805 | |
| GO:0008474 | palmitoyl-(protein) hydrolase activity | ISS | - | |
| GO:0016290 | palmitoyl-CoA hydrolase activity | IDA | 10658183 | |
| GO:0016290 | palmitoyl-CoA hydrolase activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEP | Brain (GO term level: 7) | 10992246 |
| GO:0007420 | brain development | IMP | Brain (GO term level: 7) | 7637805 |
| GO:0048666 | neuron development | TAS | neuron (GO term level: 9) | 11136716 |
| GO:0007269 | neurotransmitter secretion | IEA | neuron, axon, Synap, serotonin, Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0050803 | regulation of synapse structure and activity | NAS | neuron, Synap (GO term level: 7) | 11136716 |
| GO:0043524 | negative regulation of neuron apoptosis | IMP | neuron (GO term level: 9) | 10737604 |11020216 |
| GO:0007399 | nervous system development | IMP | neurite (GO term level: 5) | 7637805 |
| GO:0002084 | protein depalmitoylation | IDA | 10658183 |10737604 | |
| GO:0002084 | protein depalmitoylation | ISS | - | |
| GO:0006309 | DNA fragmentation during apoptosis | IDA | 15929065 | |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0016042 | lipid catabolic process | IDA | 8816748 | |
| GO:0007042 | lysosomal lumen acidification | IMP | 11722572 | |
| GO:0008306 | associative learning | IEA | - | |
| GO:0008344 | adult locomotory behavior | IEA | - | |
| GO:0007625 | grooming behavior | IEA | - | |
| GO:0007601 | visual perception | IEA | - | |
| GO:0015031 | protein transport | IMP | 10737604 | |
| GO:0050896 | response to stimulus | IEA | - | |
| GO:0051181 | cofactor transport | IMP | 16542649 | |
| GO:0051186 | cofactor metabolic process | IMP | 16542649 | |
| GO:0048260 | positive regulation of receptor-mediated endocytosis | IMP | 16542649 | |
| GO:0031579 | membrane raft organization | IMP | 15929065 | |
| GO:0030149 | sphingolipid catabolic process | TAS | 16542649 | |
| GO:0030308 | negative regulation of cell growth | IMP | 10737604 | |
| GO:0032429 | regulation of phospholipase A2 activity | IEA | - | |
| GO:0044257 | cellular protein catabolic process | IEA | - | |
| GO:0048549 | positive regulation of pinocytosis | IMP | 16542649 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0008021 | synaptic vesicle | IDA | Synap, Neurotransmitter (GO term level: 12) | 12483688 |
| GO:0030425 | dendrite | IEA | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0030424 | axon | IDA | neuron, axon, Neurotransmitter (GO term level: 6) | 12483688 |
| GO:0005794 | Golgi apparatus | IDA | 7637805 | |
| GO:0005829 | cytosol | ISS | - | |
| GO:0005576 | extracellular region | IDA | 8895569 | |
| GO:0005576 | extracellular region | ISS | - | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0005624 | membrane fraction | ISS | - | |
| GO:0005634 | nucleus | IDA | 10992246 | |
| GO:0005764 | lysosome | IDA | 8895569 |11020216 | |
| GO:0005764 | lysosome | ISS | - | |
| GO:0045121 | membrane raft | IDA | 15929065 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG DN | 45 | 29 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS UP | 79 | 46 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WANG THOC1 TARGETS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |