Gene Page: MAP2K1
Summary ?
| GeneID | 5604 |
| Symbol | MAP2K1 |
| Synonyms | CFC3|MAPKK1|MEK1|MKK1|PRKMK1 |
| Description | mitogen-activated protein kinase kinase 1 |
| Reference | MIM:176872|HGNC:HGNC:6840|HPRD:01469| |
| Gene type | protein-coding |
| Map location | 15q22.1-q22.33 |
| Pascal p-value | 0.622 |
| Sherlock p-value | 0.245 |
| Fetal beta | -2.204 |
| DMG | 1 (# studies) |
| Support | CANABINOID DOPAMINE INTRACELLULAR SIGNAL TRANSDUCTION METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAP2K1 | chr15 | 66774256 | G | T | Within intron | intronic | Schizophrenia | DNM:Awadalla_2010 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00319686 | 15 | 66680551 | MAP2K1 | 7.18E-5 | 0.897 | 0.025 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
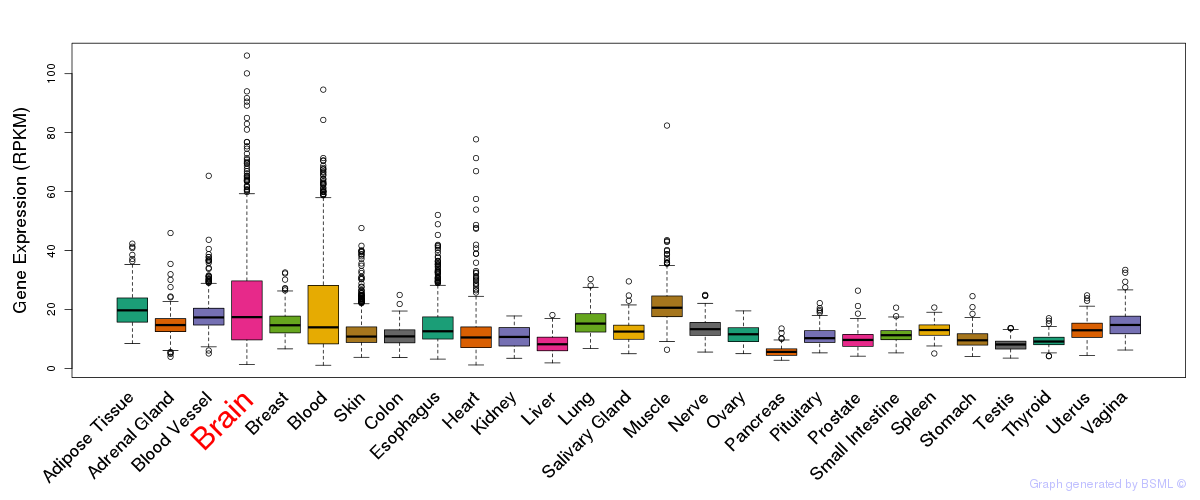
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PGD | 0.97 | 0.93 |
| RFWD2 | 0.97 | 0.94 |
| EIF4A1 | 0.96 | 0.95 |
| XRN2 | 0.96 | 0.93 |
| TRAFD1 | 0.96 | 0.91 |
| C12orf41 | 0.96 | 0.95 |
| BZW2 | 0.96 | 0.93 |
| KDM1 | 0.96 | 0.92 |
| SIAH1 | 0.96 | 0.92 |
| NARS2 | 0.96 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.76 | -0.79 |
| AF347015.27 | -0.75 | -0.90 |
| AIFM3 | -0.74 | -0.78 |
| AF347015.31 | -0.74 | -0.87 |
| MT-CO2 | -0.74 | -0.89 |
| AF347015.33 | -0.74 | -0.88 |
| MT-CYB | -0.73 | -0.89 |
| C5orf53 | -0.72 | -0.71 |
| AF347015.8 | -0.72 | -0.89 |
| AF347015.15 | -0.71 | -0.88 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004708 | MAP kinase kinase activity | TAS | 8388392 | |
| GO:0005515 | protein binding | IPI | 15299019 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030182 | neuron differentiation | IEA | neuron (GO term level: 8) | - |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0006935 | chemotaxis | TAS | 10570282 | |
| GO:0006928 | cell motion | TAS | 10912793 | |
| GO:0030216 | keratinocyte differentiation | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARAF | A-RAF | ARAF1 | PKS2 | RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | MEK1 interacts with A-Raf. | BIND | 8621729 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | MEK1 interacts with B-Raf. | BIND | 8621729 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD | 7731720 |
| CPNE1 | COPN1 | CPN1 | MGC1142 | copine I | - | HPRD | 12522145 |
| CPNE4 | COPN4 | CPN4 | MGC15604 | copine IV | - | HPRD | 12522145 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 9553107 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7969158 |
| KSR2 | FLJ25965 | kinase suppressor of ras 2 | - | HPRD,BioGRID | 12975377 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Biochemical Activity | BioGRID | 8226933 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western | BioGRID | 10969079 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | MEKK1 interacts with MEK1. | BIND | 10969079 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | MEK1 interacts with MLK3. | BIND | 15258589 |
| MAP3K4 | FLJ42439 | KIAA0213 | MAPKKK4 | MEKK4 | MTK1 | PRO0412 | mitogen-activated protein kinase kinase kinase 4 | - | HPRD,BioGRID | 9841871 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | Biochemical Activity | BioGRID | 8631303 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | MEK1 interacts with and phosphorylates ERK2. This interaction is modeled on a demonstrated interaction between human MEK1 and rat ERK2. | BIND | 11134045 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10757792 |11266467 |11279118 |11352917 |11823456 |12697810 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD | 12697810 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Affinity Capture-Western Biochemical Activity Co-localization Reconstituted Complex Two-hybrid | BioGRID | 8226933 |8626767 |9006895 |9733512 |10748187 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | MEK1 interacts with ERK1 via a conserved docking site in MEK | BIND | 11134045 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 8226933 |8626767 |10748187 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD,BioGRID | 10523642 |11044439 |
| MAPKSP1 | MAP2K1IP1 | MAPBP | MP1 | MAPK scaffold protein 1 | - | HPRD,BioGRID | 9733512 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD | 7957875 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | - | HPRD,BioGRID | 10757792 |
| PEBP4 | CORK-1 | CORK1 | GWTM1933 | MGC22776 | PRO4408 | phosphatidylethanolamine-binding protein 4 | - | HPRD,BioGRID | 15302887 |
| PEBP4 | CORK-1 | CORK1 | GWTM1933 | MGC22776 | PRO4408 | phosphatidylethanolamine-binding protein 4 | hPEBP4 interacts with MEK1. This interaction was modeled on a demonstrated interaction between human hPEBP4 and mouse MEK1. | BIND | 15302887 |
| PLAU | ATF | UPA | URK | UROKINASE | u-PA | plasminogen activator, urokinase | Phenotypic Suppression | BioGRID | 10402467 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Reconstituted Complex | BioGRID | 10757792 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf interacts with and phosphorylates Mek1 | BIND | 14724641 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | MEK1 interacts with and is phosphorylated by Raf-1. This interaction was modeled on a demonstrated interaction between human MEK1 and Raf-1 from an unspecified species. | BIND | 15866172 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | MEK1 interacts with c-Raf | BIND | 8621729 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD | 11604401 |
| ROBLD3 | ENDAP | HSPC003 | MAPBPIP | MAPKSP1AP | p14 | roadblock domain containing 3 | Affinity Capture-Western | BioGRID | 11266467 |
| RPS6KA2 | HU-2 | MAPKAPK1C | RSK | RSK3 | S6K-alpha | S6K-alpha2 | p90-RSK3 | pp90RSK3 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | - | HPRD | 8939914 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | - | HPRD | 8626767 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 8900182 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 463 | 470 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-181 | 728 | 734 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-324-3p | 440 | 446 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-330 | 458 | 464 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-34/449 | 573 | 580 | 1A,m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-503 | 464 | 470 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-96 | 131 | 137 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.