Gene Page: RAF1
Summary ?
| GeneID | 5894 |
| Symbol | RAF1 |
| Synonyms | CMD1NN|CRAF|NS5|Raf-1|c-Raf |
| Description | Raf-1 proto-oncogene, serine/threonine kinase |
| Reference | MIM:164760|HGNC:HGNC:9829|Ensembl:ENSG00000132155|HPRD:01265|Vega:OTTHUMG00000129789 |
| Gene type | protein-coding |
| Map location | 3p25 |
| Pascal p-value | 0.01 |
| Sherlock p-value | 0.43 |
| Fetal beta | 1.215 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | CANABINOID DOPAMINE METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.humanNRC G2Cdb.humanPSP Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1134 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09326546 | 3 | 12705291 | RAF1 | -0.025 | 0.25 | DMG:Nishioka_2013 | |
| cg09326546 | 3 | 12705291 | RAF1 | 9.24E-9 | -0.008 | 4.15E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | RAF1 | 5894 | 2.8E-5 | trans | ||
| rs7787830 | chr7 | 98797019 | RAF1 | 5894 | 0.05 | trans | ||
| rs16955618 | chr15 | 29937543 | RAF1 | 5894 | 4.884E-5 | trans |
Section II. Transcriptome annotation
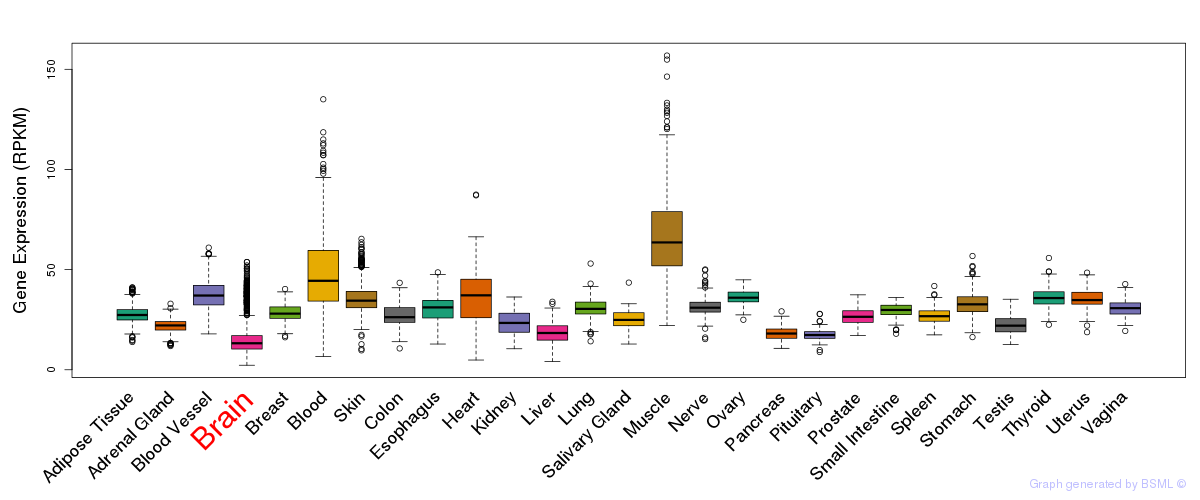
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAD50 | 0.95 | 0.95 |
| BCLAF1 | 0.94 | 0.95 |
| PRPF4B | 0.93 | 0.95 |
| DDX46 | 0.93 | 0.95 |
| NARG1 | 0.93 | 0.95 |
| ATRX | 0.93 | 0.96 |
| ZMYM2 | 0.93 | 0.93 |
| FBXO11 | 0.93 | 0.94 |
| ZNF33A | 0.93 | 0.94 |
| RBM26 | 0.93 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.73 | -0.85 |
| AF347015.31 | -0.73 | -0.85 |
| TSC22D4 | -0.73 | -0.82 |
| MT-CO2 | -0.72 | -0.85 |
| IFI27 | -0.72 | -0.85 |
| AF347015.27 | -0.71 | -0.82 |
| AIFM3 | -0.71 | -0.77 |
| ENHO | -0.70 | -0.87 |
| AF347015.33 | -0.70 | -0.81 |
| HIGD1B | -0.70 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005057 | receptor signaling protein activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 9624170 |10433554 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019992 | diacylglycerol binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | TAS | 8063729 | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0007010 | cytoskeleton organization | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 8063729 | |
| GO:0048011 | nerve growth factor receptor signaling pathway | IEA | - | |
| GO:0006915 | apoptosis | TAS | 8929532 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9069260 | |
| GO:0005741 | mitochondrial outer membrane | TAS | 8929532 | |
| GO:0005886 | plasma membrane | EXP | 9069260 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 10576742 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 8349631 |
| BAG1 | RAP46 | BCL2-associated athanogene | - | HPRD,BioGRID | 8692945 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 8910675 |8929532 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Reconstituted Complex | BioGRID | 8929532 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | B-RAF interacts with C-RAF. This interaction was modelled on a demonstrated interaction between human B-RAF and monkey C-RAF . | BIND | 15035987 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD,BioGRID | 11325826 |
| CCT3 | CCT-gamma | CCTG | PIG48 | TCP-1-gamma | TRIC5 | chaperonin containing TCP1, subunit 3 (gamma) | RAF1 (C-Raf) interacts with CCT3 (hC87). | BIND | 12620389 |
| CCT3 | CCT-gamma | CCTG | PIG48 | TCP-1-gamma | TRIC5 | chaperonin containing TCP1, subunit 3 (gamma) | C-RAF interacts with CCT3. | BIND | 12620389 |
| CCT3 | CCT-gamma | CCTG | PIG48 | TCP-1-gamma | TRIC5 | chaperonin containing TCP1, subunit 3 (gamma) | Two-hybrid | BioGRID | 12620389 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD,BioGRID | 9230211 |10373531 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD,BioGRID | 10837247 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | - | HPRD,BioGRID | 15075335 |
| CNKSR2 | CNK2 | KIAA0902 | KSR2 | connector enhancer of kinase suppressor of Ras 2 | Affinity Capture-Western | BioGRID | 14597674 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | RAF1 (C-Raf) interacts with COPS3 (SGN3). | BIND | 12620389 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | C-RAF interacts with SGN3. | BIND | 12620389 |
| CPS1 | - | carbamoyl-phosphate synthetase 1, mitochondrial | RAF1 (C-Raf) interacts with CPS1. | BIND | 12620389 |
| CPS1 | - | carbamoyl-phosphate synthetase 1, mitochondrial | C-RAF interacts with CPS1. | BIND | 12620389 |
| DMPK | DM | DM1 | DM1PK | DMK | MDPK | MT-PK | dystrophia myotonica-protein kinase | - | HPRD | 10869570 |
| EFEMP1 | DHRD | DRAD | FBLN3 | FBNL | FLJ35535 | MGC111353 | MLVT | MTLV | S1-5 | EGF-containing fibulin-like extracellular matrix protein 1 | RAF1 (C-Raf) interacts with an unspecified isoform of EFEMP1 (FBNL). | BIND | 12620389 |
| ERAS | HRAS2 | HRASP | MGC126691 | MGC126693 | ES cell expressed Ras | Affinity Capture-Western | BioGRID | 12774123 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 7517401 |
| GNB2 | - | guanine nucleotide binding protein (G protein), beta polypeptide 2 | Affinity Capture-Western | BioGRID | 7782277 |
| GNG4 | DKFZp547K1018 | FLJ23803 | FLJ34187 | guanine nucleotide binding protein (G protein), gamma 4 | - | HPRD,BioGRID | 7782277 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 9553107 |10585452 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 8530446 |8911690 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Hras interacts with Raf. This interaction was modelled on a demonstrated interaction between Hras and Raf, both from an unspecified species. | BIND | 15705808 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Raf interacts with Ras. This interaction was modeled on a demonstrated interaction between human Raf and rat Ras. | BIND | 11585923 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | RAF1 (C-Raf) interacts with HRAS (Ha-Ras). | BIND | 12620389 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Raf-1 interacts with Ras. | BIND | 15664191 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | C-RAF interacts with Ha-Ras. | BIND | 12620389 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Raf interacts with Ras. This interaction was modeled on a demonstrated interaction between Raf and an unspecified source and human Ras. | BIND | 7523381 |8307946 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7862125 |7969158 |8530446 |8628317 |9150145 |9154803 |9523700 |10369681 |10783161 |10882715 |10922060 |12379659 |12620389 |14654780 |14724584 |15031288 |16803888 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Raf-1 interacts with Hsp90. This interaction was modeled on a demonstrated interaction between human Raf-1 and monkey Hsp90. | BIND | 15618521 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 8408024 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD | 11409918 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9446616 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 8876196 |
| KRAS | C-K-RAS | K-RAS2A | K-RAS2B | K-RAS4A | K-RAS4B | KI-RAS | KRAS1 | KRAS2 | NS3 | RASK2 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | Affinity Capture-Western Two-hybrid | BioGRID | 10783161 |10882715 |
| KSR2 | FLJ25965 | kinase suppressor of ras 2 | - | HPRD | 12975377 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 8939988 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Two-hybrid | BioGRID | 10783161 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Reconstituted Complex | BioGRID | 10757792 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Raf interacts with and phosphorylates Mek1 | BIND | 14724641 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | MEK1 interacts with and is phosphorylated by Raf-1. This interaction was modeled on a demonstrated interaction between human MEK1 and Raf-1 from an unspecified species. | BIND | 15866172 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | MEK1 interacts with c-Raf | BIND | 8621729 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 11604401 |
| MAP2K2 | FLJ26075 | MAPKK2 | MEK2 | MKK2 | PRKMK2 | mitogen-activated protein kinase kinase 2 | Raf interacts with Mek2. | BIND | 14724641 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | MEKK1 interacts with Raf-1. | BIND | 10969079 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10969079 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 11427728 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western | BioGRID | 11409918 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Raf-1 interacts with ERK2. This interaction was modelled on a demonstrated interaction between human Raf-1 and ERK2 from an unspecified species. | BIND | 15664191 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Affinity Capture-Western | BioGRID | 11409918 |
| MAPK7 | BMK1 | ERK4 | ERK5 | PRKM7 | mitogen-activated protein kinase 7 | - | HPRD,BioGRID | 10531364 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD,BioGRID | 10523642 |11044439 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD | 11044439 |
| MRAS | FLJ42964 | M-RAs | R-RAS3 | RRAS3 | muscle RAS oncogene homolog | - | HPRD,BioGRID | 10498616 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 9315742 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 11005817 |
| NRAS | ALPS4 | N-ras | NRAS1 | neuroblastoma RAS viral (v-ras) oncogene homolog | Nras interacts with Raf. This interaction was modelled on a demonstrated interaction between Nras and Raf, both from an unspecified species. | BIND | 15705808 |
| NRAS | ALPS4 | N-ras | NRAS1 | neuroblastoma RAS viral (v-ras) oncogene homolog | Affinity Capture-Western | BioGRID | 10882715 |
| NUDT14 | UGPP | UGPPase | nudix (nucleoside diphosphate linked moiety X)-type motif 14 | RAF1 (C-Raf) interacts with NUDT14 (hA21). | BIND | 12620389 |
| OIP5 | 5730547N13Rik | LINT-25 | MIS18beta | Opa interacting protein 5 | Two-hybrid | BioGRID | 12620389 |
| OIP5 | 5730547N13Rik | LINT-25 | MIS18beta | Opa interacting protein 5 | C-RAF interacts with OIP5. | BIND | 12620389 |
| OIP5 | 5730547N13Rik | LINT-25 | MIS18beta | Opa interacting protein 5 | RAF1 (C-Raf) interacts with OIP5 (hC73). | BIND | 12620389 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | - | HPRD,BioGRID | 11733498 |
| PBK | FLJ14385 | Nori-3 | SPK | TOPK | PDZ binding kinase | RAF1 (C-Raf) interacts with PBK (TOPK). | BIND | 12620389 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD,BioGRID | 2475255 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | - | HPRD | 11853019 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | RKIP interacts with Raf-1. This interaction was modelled on a demonstrated interaction between RKIP from an unspecified species and Raf-1 from an unspecified species. | BIND | 14654844 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10757792 |
| PEBP4 | CORK-1 | CORK1 | GWTM1933 | MGC22776 | PRO4408 | phosphatidylethanolamine-binding protein 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15302887 |
| PEBP4 | CORK-1 | CORK1 | GWTM1933 | MGC22776 | PRO4408 | phosphatidylethanolamine-binding protein 4 | hPEBP4 interacts with Raf-1. This interaction was modeled on a demonstrated interaction between human hPEBP4 and mouse Raf-1. | BIND | 15302887 |
| PEX5L | PEX5R | PXR2 | PXR2B | peroxisomal biogenesis factor 5-like | - | HPRD | 15302887 |
| PHB | PHB1 | prohibitin | - | HPRD,BioGRID | 10523633 |
| PHB | PHB1 | prohibitin | RAF1 (c-Raf) interacts with PHB. | BIND | 16041367 |
| PHKA2 | GSD9A | MGC133071 | PHK | PYK | PYKL | XLG | XLG2 | phosphorylase kinase, alpha 2 (liver) | Two-hybrid | BioGRID | 12620389 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Raf-1 interacts with Pin1. This interaction was modelled on a demonstrated interaction between human Raf-1 and Pin1 from monkey and unspecified species. | BIND | 15664191 |
| PKM2 | CTHBP | MGC3932 | OIP3 | PK3 | PKM | TCB | THBP1 | pyruvate kinase, muscle | - | HPRD | 12620389 |
| PKM2 | CTHBP | MGC3932 | OIP3 | PK3 | PKM | TCB | THBP1 | pyruvate kinase, muscle | C-RAF interacts with PyK. | BIND | 12620389 |
| PKM2 | CTHBP | MGC3932 | OIP3 | PK3 | PKM | TCB | THBP1 | pyruvate kinase, muscle | RAF1 (C-Raf) interacts with PKM2 (PyK). | BIND | 12620389 |
| PPP2R2A | B55-ALPHA | B55A | FLJ26613 | MGC52248 | PR52A | PR55A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | RAF1 (c-Raf) interact with an unspecified isoform of PPP2R2A (PP2A-A). | BIND | 16041367 |
| PPP2R2B | B55-BETA | FLJ95686 | MGC24888 | PP2A-B55BETA | PP2A-PR55B | PP2AB-BETA | PP2APR55-BETA | PR2AB-BETA | PR2AB55-BETA | PR2APR55-BETA | PR52B | PR55-BETA | SCA12 | protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform | RAF1 (c-Raf) interacts with an unspecified isoform of PPP2R2B (PP2A-B). | BIND | 16041367 |
| PPP2R2C | B55-GAMMA | IMYPNO | IMYPNO1 | MGC33570 | PR52 | PR55G | protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | RAF1 (c-Raf) interacts with an unspecified isoform of PPP2R2C (PP2A-C). | BIND | 16041367 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD,BioGRID | 11350735 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 10620507 |
| PRKG1 | CGKI | DKFZp686K042 | FLJ36117 | MGC71944 | PGK | PKG | PRKG1B | PRKGR1B | cGKI-BETA | cGKI-alpha | protein kinase, cGMP-dependent, type I | Affinity Capture-Western | BioGRID | 12237340 |
| PRPF6 | ANT-1 | C20orf14 | TOM | U5-102K | hPrp6 | PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 10848612 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 7791872 |7862125 |9867809 |10454553 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | - | HPRD | 7791872|9867809|10454553 |
| RAP2A | K-REV | KREV | RAP2 | RbBP-30 | RAP2A, member of RAS oncogene family | Raf-1 interacts with Rap2. This interaction was modeled on a demonstrated interaction between Raf-1 from unspecified species and human Rap2. | BIND | 15752761 |
| RASD2 | MGC:4834 | Rhes | TEM2 | RASD family, member 2 | Reconstituted Complex | BioGRID | 14724584 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 9819434 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Raf-1 interacts with Rb. | BIND | 9819434 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | Raf-1 interacts with p130. | BIND | 9819434 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | - | HPRD,BioGRID | 9819434 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | - | HPRD,BioGRID | 9001246 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | RAF interacts with Rheb. | BIND | 15854902 |
| RP6-213H19.1 | MASK | MST4 | serine/threonine protein kinase MST4 | - | HPRD,BioGRID | 11306563 |
| RRAS | - | related RAS viral (r-ras) oncogene homolog | Two-hybrid | BioGRID | 10848612 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | RAF1 (C-Raf) interacts with RRAS2 (R-Ras). | BIND | 12620389 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | Reconstituted Complex Two-hybrid | BioGRID | 10557073 |10783161 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD | 10064593 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SHOC2 | FLJ60412 | KIAA0862 | SIAA0862 | SOC-2 | SOC2 | SUR-8 | SUR8 | soc-2 suppressor of clear homolog (C. elegans) | - | HPRD,BioGRID | 10783161 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | Affinity Capture-Western | BioGRID | 12717443 |
| SPRY4 | - | sprouty homolog 4 (Drosophila) | - | HPRD,BioGRID | 12717443 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 7517401 |
| STK3 | FLJ90748 | KRS1 | MST2 | serine/threonine kinase 3 (STE20 homolog, yeast) | Raf-1 interacts with MST2. This interaction was modeled on a demonstrated interaction between human Raf-1 and monkey MST2. | BIND | 15618521 |
| TIMM50 | MGC102733 | TIM50 | TIM50L | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | C-RAF interacts with TIMM50. | BIND | 12620389 |
| TIMM50 | MGC102733 | TIM50 | TIM50L | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | RAF1 (C-Raf) interacts with TIMM50 (hC22). | BIND | 12620389 |
| TSC22D3 | DIP | DKFZp313A1123 | DSIPI | GILZ | TSC-22R | hDIP | TSC22 domain family, member 3 | - | HPRD,BioGRID | 12391160 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | Biochemical Activity | BioGRID | 11676916 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 8900182 |
| VDAC1 | MGC111064 | PORIN | PORIN-31-HL | voltage-dependent anion channel 1 | Affinity Capture-Western | BioGRID | 12079506 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | C-RAF interacts with 14-3-3-beta. | BIND | 12620389 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD | 8702721 |12360521 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | RAF1 (C-Raf) interacts with YWHAB (14-3-3-beta). | BIND | 12620389 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8702721 |10620507 |10848612 |12360521 |12620389 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 7644510 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 14-3-3-epsilon interacts with Raf-1. | BIND | 7644510 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 8702721 |10433554 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10433554 |10620507 |17353931 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD,BioGRID | 8702721 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8663231 |10620507 |10757792 |17353931 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Raf-1 interacts with 14-3-3 zeta. This interaction was modelled on a demonstrated interaction between human Raf-1 and murine 14-3-3-zeta. | BIND | 7559537 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 7628630 |9261098 |10620507 |10887173 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 7628630 |7939632 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 139 | 145 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-19 | 461 | 468 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-331 | 27 | 34 | 1A,m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-503 | 108 | 114 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-7 | 674 | 681 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.