Gene Page: RANBP2
Summary ?
| GeneID | 5903 |
| Symbol | RANBP2 |
| Synonyms | ADANE|ANE1|IIAE3|NUP358|TRP1|TRP2 |
| Description | RAN binding protein 2 |
| Reference | MIM:601181|HGNC:HGNC:9848|Ensembl:ENSG00000153201|HPRD:03111|Vega:OTTHUMG00000130981 |
| Gene type | protein-coding |
| Map location | 2q12.3 |
| Pascal p-value | 0.357 |
| Sherlock p-value | 0.176 |
| Fetal beta | 0.227 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.024 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08028004 | 2 | 109336023 | RANBP2 | -0.034 | 0.25 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13023481 | chr2 | 213839772 | RANBP2 | 5903 | 0.08 | trans |
Section II. Transcriptome annotation
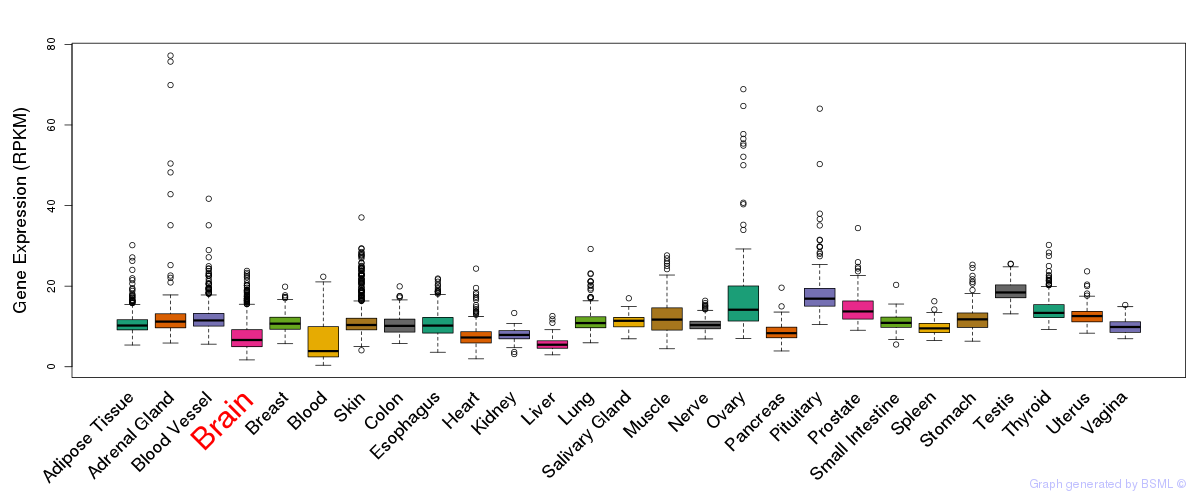
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TEX10 | 0.95 | 0.92 |
| KLHL7 | 0.95 | 0.92 |
| DDX5 | 0.95 | 0.88 |
| HSF2 | 0.95 | 0.92 |
| XRN2 | 0.94 | 0.89 |
| CEP57 | 0.94 | 0.90 |
| NOL11 | 0.94 | 0.91 |
| POLR2B | 0.94 | 0.91 |
| ZNF195 | 0.94 | 0.89 |
| DHX9 | 0.94 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.75 | -0.82 |
| AIFM3 | -0.73 | -0.81 |
| MT-CO2 | -0.72 | -0.87 |
| AF347015.33 | -0.72 | -0.87 |
| AF347015.31 | -0.72 | -0.85 |
| AF347015.27 | -0.72 | -0.86 |
| MT-CYB | -0.72 | -0.88 |
| HEPN1 | -0.71 | -0.81 |
| TSC22D4 | -0.70 | -0.81 |
| AF347015.8 | -0.70 | -0.87 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 16332688 |17353931 |18394993 | |
| GO:0016853 | isomerase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008536 | Ran GTPase binding | TAS | 7603572 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006457 | protein folding | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006606 | protein import into nucleus | TAS | 7603572 | |
| GO:0051028 | mRNA transport | IEA | - | |
| GO:0015031 | protein transport | IEA | - | |
| GO:0046907 | intracellular transport | IEA | - | |
| GO:0065002 | intracellular protein transmembrane transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 12228227 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005643 | nuclear pore | TAS | 7603572 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| IPO5 | DKFZp686O1576 | FLJ43041 | IMB3 | KPNB3 | MGC2068 | RANBP5 | importin 5 | - | HPRD | 9114010 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | Reconstituted Complex | BioGRID | 10473610 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | - | HPRD,BioGRID | 10473610 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | - | HPRD | 11266456 |
| OPN1LW | CBBM | CBP | RCP | opsin 1 (cone pigments), long-wave-sensitive | - | HPRD,BioGRID | 8857542 |
| OPN1MW | CBBM | CBD | GCP | MGC176615 | OPN1MW1 | opsin 1 (cone pigments), medium-wave-sensitive | - | HPRD,BioGRID | 8857542 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | - | HPRD,BioGRID | 10318915 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | - | HPRD | 9019411 |11854305 |12192048 |
| SAE1 | AOS1 | FLJ3091 | HSPC140 | SUA1 | SUMO1 activating enzyme subunit 1 | Biochemical Activity | BioGRID | 11792325 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | Biochemical Activity | BioGRID | 18691969 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Reconstituted Complex | BioGRID | 12192048 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | SUMO-1 interacts with RanBP2. | BIND | 15608651 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD | 9144189 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | Biochemical Activity | BioGRID | 11792325 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Ubc9 interacts with RanBP2. | BIND | 15608651 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | RanBP2 interacts with Ubc9. | BIND | 15378033 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 12192048 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD | 10601307 |
| XPOT | XPO3 | exportin, tRNA (nuclear export receptor for tRNAs) | - | HPRD | 12138183 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE TRANSPORT | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS B UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 700 | 707 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-133 | 378 | 384 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-137 | 270 | 277 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-142-5p | 819 | 825 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-153 | 306 | 312 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-196 | 699 | 705 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-199 | 194 | 200 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-217 | 835 | 841 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-221/222 | 860 | 866 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-377 | 175 | 181 | 1A | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-448 | 305 | 312 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-496 | 114 | 120 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 504 | 510 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.