Gene Page: SMARCC2
Summary ?
| GeneID | 6601 |
| Symbol | SMARCC2 |
| Synonyms | BAF170|CRACC2|Rsc8 |
| Description | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| Reference | MIM:601734|HGNC:HGNC:11105|Ensembl:ENSG00000139613|HPRD:03437|Vega:OTTHUMG00000170288 |
| Gene type | protein-coding |
| Map location | 12q13.2 |
| Pascal p-value | 0.355 |
| Sherlock p-value | 0.08 |
| TADA p-value | 0.003 |
| Fetal beta | -0.528 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0178 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMARCC2 | chr12 | 56566768..56566770 | CTT | C | NM_001130420 NM_003075 NM_139067 | . . . | frameshift intronic frameshift | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01982788 | 12 | 56556339 | SMARCC2 | 4.872E-4 | 0.342 | 0.047 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs163562 | chr3 | 3085003 | SMARCC2 | 6601 | 0.17 | trans |
Section II. Transcriptome annotation
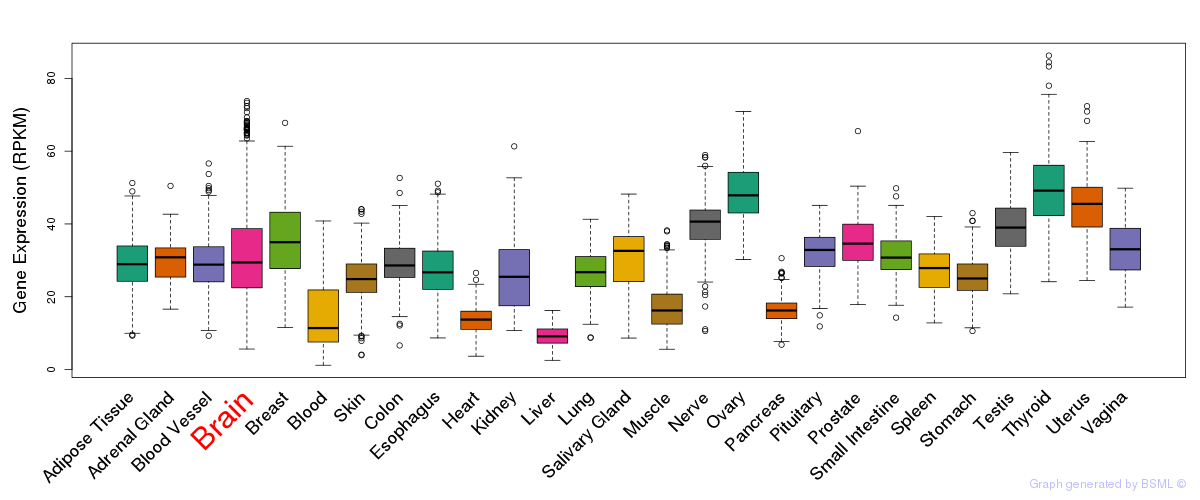
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SSPN | 0.84 | 0.84 |
| PDLIM5 | 0.84 | 0.87 |
| AQP4 | 0.81 | 0.77 |
| TIMP3 | 0.81 | 0.83 |
| MTM1 | 0.80 | 0.79 |
| EDNRB | 0.79 | 0.69 |
| FIBIN | 0.77 | 0.77 |
| MYOM1 | 0.76 | 0.79 |
| TMEM47 | 0.76 | 0.78 |
| SPATA6 | 0.74 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.40 | -0.48 |
| PLEKHO1 | -0.39 | -0.44 |
| COMMD3 | -0.38 | -0.49 |
| SLA | -0.37 | -0.25 |
| IER5L | -0.37 | -0.33 |
| MPPED1 | -0.37 | -0.32 |
| ISLR2 | -0.37 | -0.27 |
| AC011491.1 | -0.37 | -0.43 |
| AC132872.1 | -0.36 | -0.50 |
| SNHG12 | -0.36 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | TAS | 8804307 | |
| GO:0008022 | protein C-terminus binding | IPI | 12917342 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006333 | chromatin assembly or disassembly | IEA | - | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 8804307 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006338 | chromatin remodeling | IDA | 10078207 |11018012 | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 12192000 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | IDA | 11018012 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000785 | chromatin | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | TAS | 8804307 | |
| GO:0017053 | transcriptional repressor complex | IPI | 12192000 | |
| GO:0016514 | SWI/SNF complex | IDA | 10078207 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | Affinity Capture-Western | BioGRID | 9845365 |
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | Two-hybrid | BioGRID | 16169070 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD,BioGRID | 12837248 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Co-purification | BioGRID | 10943845 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Co-fractionation | BioGRID | 9710619 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Reconstituted Complex | BioGRID | 11018012 |
| GRM2 | GLUR2 | GPRC1B | MGLUR2 | mGlu2 | glutamate receptor, metabotropic 2 | BAF170 interacts with mGluR2 promoter. | BIND | 15035981 |
| HSP90B1 | ECGP | GP96 | GRP94 | TRA1 | heat shock protein 90kDa beta (Grp94), member 1 | Affinity Capture-Western | BioGRID | 11726552 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | Affinity Capture-MS | BioGRID | 11784859 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | Two-hybrid | BioGRID | 16169070 |
| KLF1 | EKLF | Kruppel-like factor 1 (erythroid) | Reconstituted Complex | BioGRID | 11018012 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 14743216 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Co-purification | BioGRID | 11013263 |
| PEX14 | MGC12767 | NAPP2 | Pex14p | dJ734G22.2 | peroxisomal biogenesis factor 14 | Two-hybrid | BioGRID | 16169070 |
| PHYHIP | DYRK1AP3 | KIAA0273 | PAHX-AP | PAHXAP1 | phytanoyl-CoA 2-hydroxylase interacting protein | Two-hybrid | BioGRID | 16169070 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 9710619 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Two-hybrid | BioGRID | 16169070 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | - | HPRD,BioGRID | 14743216 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Co-purification | BioGRID | 8895581 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |11726552 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Co-purification | BioGRID | 8895581 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Affinity Capture-Western | BioGRID | 12917342 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | Affinity Capture-Western | BioGRID | 9845365 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 11018012 |
| SRGAP3 | ARHGAP14 | KIAA0411 | MEGAP | SRGAP2 | WRP | SLIT-ROBO Rho GTPase activating protein 3 | - | HPRD | 12368262 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | Affinity Capture-Western | BioGRID | 12244326 |
| STMN2 | SCG10 | SCGN10 | SGC10 | stathmin-like 2 | BAF170 interacts with SCG10 promoter. | BIND | 15035981 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-150 | 220 | 226 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.