Gene Page: SMO
Summary ?
| GeneID | 6608 |
| Symbol | SMO |
| Synonyms | FZD11|Gx|SMOH |
| Description | smoothened, frizzled class receptor |
| Reference | MIM:601500|HGNC:HGNC:11119|Ensembl:ENSG00000128602|HPRD:03294|Vega:OTTHUMG00000158421 |
| Gene type | protein-coding |
| Map location | 7q32.3 |
| Pascal p-value | 0.744 |
| Fetal beta | 1.173 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19015909 | 7 | 128828512 | SMO | 6.97E-8 | 0.006 | 1.69E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11080561 | chr18 | 12257229 | SMO | 6608 | 0 | trans |
Section II. Transcriptome annotation
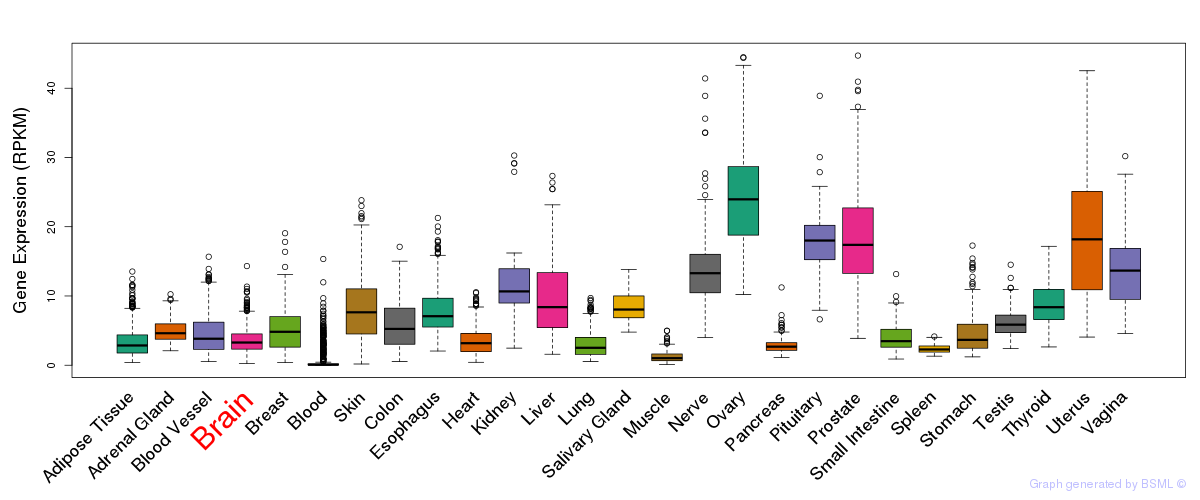
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CD99L2 | 0.88 | 0.86 |
| MAP7D2 | 0.87 | 0.87 |
| PLEKHB2 | 0.87 | 0.86 |
| AARS | 0.86 | 0.85 |
| DDHD2 | 0.86 | 0.85 |
| MAP1A | 0.86 | 0.84 |
| RNF157 | 0.86 | 0.84 |
| NSF | 0.86 | 0.86 |
| EHD3 | 0.86 | 0.85 |
| ATP6V0A1 | 0.86 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf61 | -0.42 | -0.43 |
| RPL35 | -0.41 | -0.47 |
| IL32 | -0.41 | -0.38 |
| SIGIRR | -0.41 | -0.48 |
| CLEC3B | -0.41 | -0.47 |
| BCL7C | -0.41 | -0.51 |
| FAM159B | -0.40 | -0.60 |
| AC006276.2 | -0.40 | -0.41 |
| GNG11 | -0.40 | -0.33 |
| RAMP2 | -0.40 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 9422511 | |
| GO:0004930 | G-protein coupled receptor activity | IEA | - | |
| GO:0005515 | protein binding | TAS | 8906787 | |
| GO:0004926 | non-G-protein coupled 7TM receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0021953 | central nervous system neuron differentiation | IEA | neuron (GO term level: 9) | - |
| GO:0001755 | neural crest cell migration | IEA | - | |
| GO:0001570 | vasculogenesis | IEA | - | |
| GO:0001649 | osteoblast differentiation | IEA | - | |
| GO:0002052 | positive regulation of neuroblast proliferation | IEA | - | |
| GO:0003007 | heart morphogenesis | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007368 | determination of left/right symmetry | IEA | - | |
| GO:0009953 | dorsal/ventral pattern formation | IEA | - | |
| GO:0048468 | cell development | IEA | - | |
| GO:0007389 | pattern specification process | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0021938 | smoothened signaling pathway involved in regulation of granule cell precursor cell proliferation | IEA | - | |
| GO:0043392 | negative regulation of DNA binding | IEA | - | |
| GO:0021696 | cerebellar cortex morphogenesis | IEA | - | |
| GO:0048589 | developmental growth | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | 9422511 | |
| GO:0005929 | cilium | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG 2PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES DN | 34 | 26 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C5 | 21 | 11 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP | 49 | 32 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 1HR DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION TOP20 DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-381 | 1086 | 1092 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.