Gene Page: SP1
Summary ?
| GeneID | 6667 |
| Symbol | SP1 |
| Synonyms | - |
| Description | Sp1 transcription factor |
| Reference | MIM:189906|HGNC:HGNC:11205|Ensembl:ENSG00000185591|HPRD:01796|Vega:OTTHUMG00000170047 |
| Gene type | protein-coding |
| Map location | 12q13.1 |
| Pascal p-value | 0.043 |
| Sherlock p-value | 0.392 |
| Fetal beta | 0.532 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0366 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | SP1 | 6667 | 0.05 | trans |
Section II. Transcriptome annotation
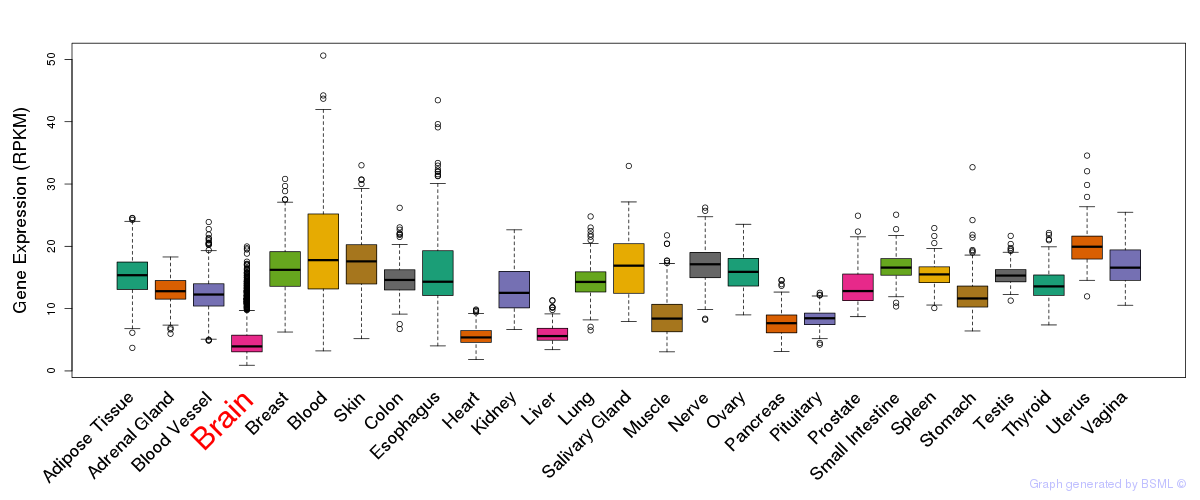
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003690 | double-stranded DNA binding | IEA | - | |
| GO:0003700 | transcription factor activity | IDA | 12560508 | |
| GO:0003702 | RNA polymerase II transcription factor activity | NAS | 3139301 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008022 | protein C-terminus binding | IPI | 10816420 | |
| GO:0016563 | transcription activator activity | IDA | 14580349 | |
| GO:0042803 | protein homodimerization activity | IDA | 10393239 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IDA | 12169688 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006350 | transcription | IEA | - | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | IDA | 14580349 | |
| GO:0045817 | positive regulation of transcription from RNA polymerase II promoter, global | IDA | 12169688 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IC | 3139301 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AATF | CHE-1 | CHE1 | DED | apoptosis antagonizing transcription factor | - | HPRD,BioGRID | 12847090 |
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 10471301 |12612060 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western | BioGRID | 15661808 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Reconstituted Complex | BioGRID | 10471301 |
| ATF7IP | FLJ10139 | FLJ10688 | MCAF | p621 | activating transcription factor 7 interacting protein | MCAF1 interacts with Sp1. | BIND | 15691849 |
| ATF7IP2 | FLJ12668 | MCAF2 | activating transcription factor 7 interacting protein 2 | MCAF2 interacts with Sp1. | BIND | 15691849 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | - | HPRD | 12004059 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 10720440 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with Sp1. | BIND | 12706836 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western | BioGRID | 12706836 |
| C14orf106 | FLJ11186 | HSA242977 | KIAA1903 | MIS18BP1 | chromosome 14 open reading frame 106 | - | HPRD | 10976766 |
| CD2 | SRBC | T11 | CD2 molecule | - | HPRD | 7933095 |
| CD209 | CDSIGN | CLEC4L | DC-SIGN | DC-SIGN1 | MGC129965 | CD209 molecule | Sp1 interacts with the CD209 promoter. | BIND | 15838506 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | Sp1 interacts with the cdc25c promoter. | BIND | 15574328 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | SP1 interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Sp1 interacts with CDKN1A promoter. | BIND | 15780936 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 12847250 |
| CLCN1 | CLC1 | MGC138361 | MGC142055 | chloride channel 1, skeletal muscle | p-CLCN1 interacts with Sp1. | BIND | 14657503 |
| DMPK | DM | DM1 | DM1PK | DMK | MDPK | MT-PK | dystrophia myotonica-protein kinase | Mutant DMPK mRNA interacts with Sp1. | BIND | 14657503 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | - | HPRD,BioGRID | 10547281 |
| E2F2 | E2F-2 | E2F transcription factor 2 | - | HPRD | 8657141 |8657142 |9694791 |
| E2F3 | DKFZp686C18211 | E2F-3 | KIAA0075 | MGC104598 | E2F transcription factor 3 | - | HPRD | 8657141 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | Egr1 interacts with Sp1.This interaction was modelled on a demonstrated interaction between human Egr1 and mouse Sp1. | BIND | 12569082 |
| ELF1 | - | E74-like factor 1 (ets domain transcription factor) | - | HPRD,BioGRID | 10976766 |
| GABPA | E4TF1-60 | E4TF1A | NFT2 | NRF2 | NRF2A | GA binding protein transcription factor, alpha subunit 60kDa | - | HPRD,BioGRID | 11237613 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Reconstituted Complex | BioGRID | 12239153 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 14970236 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | - | HPRD,BioGRID | 10976766 |12670868 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western | BioGRID | 11896613 |12091390 |12176973 |12847090 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 11896613 |11909966 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 12151407 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | Sp1 interacts with HIF-1alpha. | BIND | 15780936 |
| HMGA1 | HMG-R | HMGA1A | HMGIY | MGC12816 | MGC4242 | MGC4854 | high mobility group AT-hook 1 | - | HPRD,BioGRID | 12665574 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 10976766 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 11839795 |
| HTT | HD | IT15 | huntingtin | Huntingtin interacts with Sp1. This interaction was modelled on a demonstrated interaction between Huntingtin from human and Sp1 from rat. | BIND | 11839795 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 10506225 |
| KLF10 | EGRA | TIEG | TIEG1 | Kruppel-like factor 10 | - | HPRD,BioGRID | 10976766 |
| KLF4 | EZF | GKLF | Kruppel-like factor 4 (gut) | - | HPRD,BioGRID | 9651398 |12034813 |
| KLF6 | BCD1 | COPEB | CPBP | DKFZp686N0199 | GBF | PAC1 | ST12 | ZF9 | Kruppel-like factor 6 | - | HPRD,BioGRID | 12433697 |
| LDB1 | CLIM2 | NLI | LIM domain binding 1 | Reconstituted Complex | BioGRID | 12239153 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | Reconstituted Complex | BioGRID | 12239153 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | SP1 interacts with the MDM2 intronic promoter. | BIND | 15550242 |
| MEF2C | - | myocyte enhancer factor 2C | - | HPRD,BioGRID | 9748305 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | - | HPRD,BioGRID | 12213324 |
| MIER1 | DKFZp781G0451 | ER1 | KIAA1610 | MGC131940 | MGC150640 | MGC150641 | MI-ER1 | RP5-944N15.1 | hMI-ER1 | mesoderm induction early response 1 homolog (Xenopus laevis) | - | HPRD | 15117948 |
| MSX1 | HOX7 | HYD1 | msh homeobox 1 | - | HPRD,BioGRID | 10215616 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Myc interacts with Sp1. This interaction was modeled on a demonstrated interaction between mouse Myc and human Sp-1. | BIND | 11274368 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Sp1 interacts with Myc. | BIND | 15780936 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 11274368 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 10082523 |
| MYOG | MYF4 | MYOGENIN | bHLHc3 | myogenin (myogenic factor 4) | - | HPRD,BioGRID | 10082523 |
| NCL | C23 | FLJ45706 | nucleolin | Affinity Capture-Western | BioGRID | 14623863 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | Reconstituted Complex | BioGRID | 7933095 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | Reconstituted Complex | BioGRID | 7933095 |
| NOS3 | ECNOS | eNOS | nitric oxide synthase 3 (endothelial cell) | - | HPRD | 7541039 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD,BioGRID | 9388268 |
| NR5A1 | AD4BP | ELP | FTZ1 | FTZF1 | SF-1 | SF1 | nuclear receptor subfamily 5, group A, member 1 | - | HPRD,BioGRID | 10919277 |
| PGR | NR3C3 | PR | progesterone receptor | Sp1 interacts with the PgRA promoter. | BIND | 15019994 |
| PGR | NR3C3 | PR | progesterone receptor | Sp1 interacts with the PgRB promoter. | BIND | 15019994 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 9819401 |
| POGZ | KIAA0461 | MGC71543 | SUHW5 | ZNF280E | ZNF635 | ZNF635m | pogo transposable element with ZNF domain | - | HPRD,BioGRID | 10976766 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD,BioGRID | 8668525 |
| PPP1R13L | IASPP | NKIP1 | RAI | protein phosphatase 1, regulatory (inhibitor) subunit 13 like | - | HPRD,BioGRID | 12134007 |
| PSIP1 | DFS70 | LEDGF | MGC74712 | PAIP | PSIP2 | p52 | p75 | PC4 and SFRS1 interacting protein 1 | Reconstituted Complex | BioGRID | 9885563 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | - | HPRD,BioGRID | 10816420 |
| PURA | PUR-ALPHA | PUR1 | PURALPHA | purine-rich element binding protein A | - | HPRD,BioGRID | 10457364 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 10361124 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD | 7732011 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western | BioGRID | 11896613 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | - | HPRD,BioGRID | 12091390 |12943729 |
| RECQL4 | RECQ4 | RecQ protein-like 4 | SP1 interacts with the RECQL4 (RECQ4) promoter. | BIND | 15674334 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 7933095 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD | 7933095|12055073 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 7933095 |12055073 |
| RNASEN | DROSHA | ETOHI2 | HSA242976 | RANSE3L | RN3 | RNASE3L | ribonuclease type III, nuclear | - | HPRD | 10976766 |
| RNF4 | RES4-26 | SNURF | ring finger protein 4 | Affinity Capture-Western | BioGRID | 10617653 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex | BioGRID | 10361124 |
| SF3A1 | PRP21 | PRPF21 | SAP114 | SF3A120 | splicing factor 3a, subunit 1, 120kDa | - | HPRD | 10976766 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 7933095 |
| SKIV2L2 | Dob1 | KIAA0052 | MGC142069 | Mtr4 | fSAP118 | superkiller viralicidic activity 2-like 2 (S. cerevisiae) | - | HPRD | 14676314 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11114293 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Affinity Capture-Western | BioGRID | 11114293 |11432852 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD | 11432852 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Reconstituted Complex | BioGRID | 11018012 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Reconstituted Complex | BioGRID | 11018012 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Reconstituted Complex | BioGRID | 11018012 |
| SP3 | DKFZp686O1631 | SPR-2 | Sp3 transcription factor | - | HPRD | 9329821 |
| SP4 | HF1B | MGC130008 | MGC130009 | SPR-1 | Sp4 transcription factor | - | HPRD,BioGRID | 7559627 |
| SREBF1 | SREBP-1c | SREBP1 | bHLHd1 | sterol regulatory element binding transcription factor 1 | - | HPRD,BioGRID | 10224053 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | - | HPRD,BioGRID | 10976766 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | Reconstituted Complex | BioGRID | 10082523 |
| SUB1 | MGC102747 | P15 | PC4 | p14 | SUB1 homolog (S. cerevisiae) | Reconstituted Complex | BioGRID | 9885563 |
| TAF4 | FLJ41943 | TAF2C | TAF2C1 | TAF4A | TAFII130 | TAFII135 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | - | HPRD,BioGRID | 9742090 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | - | HPRD,BioGRID | 12239153 |
| TNFSF10 | APO2L | Apo-2L | CD253 | TL2 | TRAIL | tumor necrosis factor (ligand) superfamily, member 10 | SP1 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with Sp1. | BIND | 15574328 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with Sp1. | BIND | 9492043 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | TP53 (p53) interacts with Sp1. | BIND | 9685344 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | TP53 (p53) interacts with SP1. | BIND | 15674334 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | Affinity Capture-Western | BioGRID | 10224053 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | YY1 interacts with Sp1. | BIND | 8327494 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD | 12004059 |
| ZBTB7A | DKFZp547O146 | FBI-1 | FBI1 | LRF | MGC99631 | ZBTB7 | ZNF857A | pokemon | zinc finger and BTB domain containing 7A | - | HPRD | 12004059 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 524 | 530 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 524 | 530 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 3141 | 3147 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 4516 | 4522 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-135 | 344 | 350 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-149 | 3809 | 3815 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-193 | 3811 | 3817 | 1A | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-214 | 3918 | 3924 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-218 | 1405 | 1411 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-22 | 4733 | 4739 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-24 | 542 | 548 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-25/32/92/363/367 | 1779 | 1785 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 1663 | 1669 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-29 | 3168 | 3174 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU | ||||
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-326 | 17 | 23 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-33 | 1580 | 1586 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-330 | 4035 | 4041 | 1A | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-335 | 4579 | 4586 | 1A,m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-361 | 3921 | 3927 | 1A | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC | ||||
| miR-369-3p | 412 | 418 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 412 | 418 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-375 | 1434 | 1440 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-376 | 3413 | 3419 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-378 | 2297 | 2303 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU | ||||
| miR-381 | 3354 | 3360 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-410 | 414 | 420 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-485-3p | 5008 | 5014 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-493-5p | 5135 | 5141 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 4906 | 4912 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-7 | 1152 | 1159 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.