Gene Page: MAP3K7
Summary ?
| GeneID | 6885 |
| Symbol | MAP3K7 |
| Synonyms | MEKK7|TAK1|TGF1a |
| Description | mitogen-activated protein kinase kinase kinase 7 |
| Reference | MIM:602614|HGNC:HGNC:6859|Ensembl:ENSG00000135341|HPRD:04011|Vega:OTTHUMG00000015217 |
| Gene type | protein-coding |
| Map location | 6q15 |
| Pascal p-value | 0.066 |
| Sherlock p-value | 0.221 |
| Fetal beta | -0.352 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0024 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01087382 | 6 | 91297190 | MAP3K7 | 9E-8 | -0.019 | 2.06E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
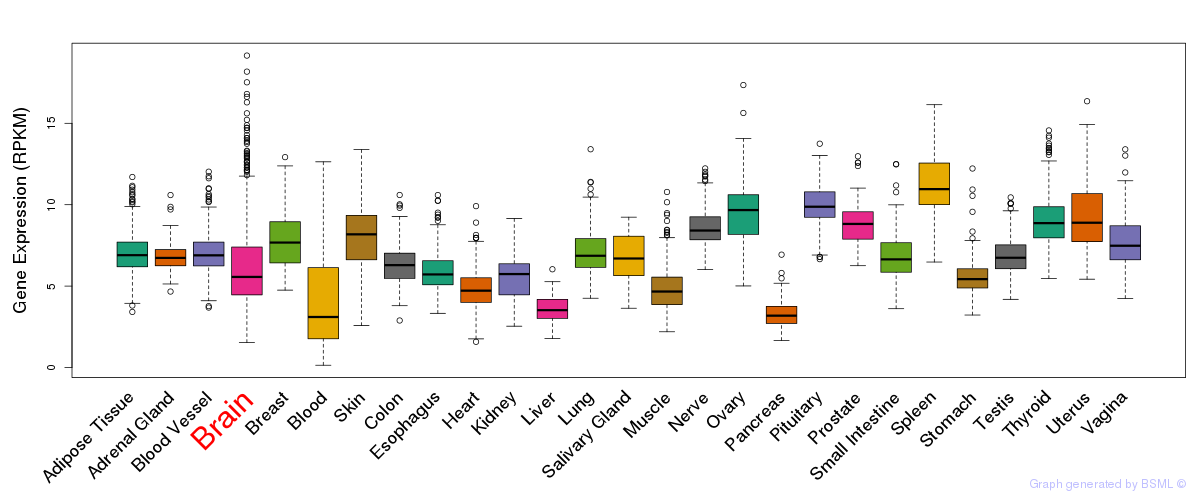
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZBTB10 | 0.89 | 0.93 |
| SPIRE1 | 0.89 | 0.94 |
| IL17RD | 0.89 | 0.88 |
| MED13 | 0.88 | 0.93 |
| RCOR1 | 0.88 | 0.91 |
| ZNF192 | 0.87 | 0.88 |
| CCNG2 | 0.87 | 0.90 |
| BMPR1A | 0.87 | 0.90 |
| RBM12 | 0.87 | 0.92 |
| RBM15 | 0.87 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AIFM3 | -0.65 | -0.74 |
| FXYD1 | -0.63 | -0.84 |
| S100B | -0.63 | -0.80 |
| AF347015.31 | -0.63 | -0.82 |
| HLA-F | -0.63 | -0.70 |
| C5orf53 | -0.63 | -0.69 |
| MT-CO2 | -0.62 | -0.83 |
| TSC22D4 | -0.62 | -0.74 |
| AF347015.27 | -0.62 | -0.79 |
| AF347015.33 | -0.62 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004708 | MAP kinase kinase activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15075345 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004709 | MAP kinase kinase kinase activity | TAS | 9466656 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000165 | MAPKKK cascade | IEA | - | |
| GO:0001525 | angiogenesis | IEA | - | |
| GO:0002726 | positive regulation of T cell cytokine production | IMP | 15125833 | |
| GO:0001841 | neural tube formation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007252 | I-kappaB phosphorylation | IEA | - | |
| GO:0007250 | activation of NF-kappaB-inducing kinase activity | IMP | 15125833 | |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | IEA | - | |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | TAS | 9466656 | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0050852 | T cell receptor signaling pathway | NAS | 15125833 | |
| GO:0050870 | positive regulation of T cell activation | IC | 15125833 | |
| GO:0032743 | positive regulation of interleukin-2 production | IMP | 15125833 | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEA | - | |
| GO:0046330 | positive regulation of JNK cascade | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9162092 |9744859 |10066798 |12609980 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | - | HPRD | 14743216 |
| CAD | - | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | - | HPRD | 14743216 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 14743216 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | - | HPRD | 14743216 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 10187861 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD | 14743216 |
| DARS | DKFZp781B11202 | MGC111579 | aspartyl-tRNA synthetase | - | HPRD | 14743216 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | - | HPRD | 14743216 |
| DNAJA1 | DJ-2 | DjA1 | HDJ2 | HSDJ | HSJ2 | HSPF4 | hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 | - | HPRD | 14743216 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with MAP3K7 promoter. | BIND | 15782160 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | - | HPRD | 14743216 |
| ENO1 | ENO1L1 | MBP-1 | MPB1 | NNE | PPH | enolase 1, (alpha) | - | HPRD | 14743216 |
| EPRS | DKFZp313B047 | EARS | GLUPRORS | PARS | PIG32 | QARS | QPRS | glutamyl-prolyl-tRNA synthetase | - | HPRD | 14743216 |
| FKBP5 | FKBP51 | FKBP54 | MGC111006 | P54 | PPIase | Ptg-10 | FK506 binding protein 5 | - | HPRD | 14743216 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 14743216 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 11397816 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD,BioGRID | 11397816 |
| HIST1H4I | H4/m | H4FM | H4M | histone cluster 1, H4i | - | HPRD | 14743216 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 14743216 |
| HSP90AB1 | D6S182 | FLJ26984 | HSP90-BETA | HSP90B | HSPC2 | HSPCB | heat shock protein 90kDa alpha (cytosolic), class B member 1 | - | HPRD | 14743216 |
| HSPA1B | FLJ54328 | HSP70-1B | HSP70-2 | HSPA1A | heat shock 70kDa protein 1B | - | HPRD | 14743216 |
| HSPA1L | HSP70-1L | HSP70-HOM | HSP70T | hum70t | heat shock 70kDa protein 1-like | - | HPRD | 14743216 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD | 14743216 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| HSPA9 | CSA | GRP75 | HSPA9B | MGC4500 | MOT | MOT2 | MTHSP75 | PBP74 | mot-2 | heat shock 70kDa protein 9 (mortalin) | - | HPRD | 14743216 |
| IARS | FLJ20736 | IARS1 | ILRS | PRO0785 | isoleucyl-tRNA synthetase | - | HPRD | 14743216 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD | 12556533 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | - | HPRD,BioGRID | 11460167 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 10094049 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 10921914 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | Reconstituted Complex | BioGRID | 9556573 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | - | HPRD,BioGRID | 10838074 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | An unspecified isoform of TAK1 interacts with TAB1. This interaction was modeled on a demonstrated interaction between TAK1 from an unspecified species and human TAB1. | BIND | 9878061 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | - | HPRD | 10838074|14743216 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD,BioGRID | 11259596 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 11259596 |14743216 |
| MAP3K7IP3 | MGC45404 | NAP1 | TAB3 | mitogen-activated protein kinase kinase kinase 7 interacting protein 3 | Affinity Capture-Western | BioGRID | 14633987 |14670075 |
| MAP4K1 | HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 | - | HPRD,BioGRID | 10224067 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | - | HPRD,BioGRID | 10807933 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD | 11865055 |
| NAIP | BIRC1 | FLJ18088 | FLJ42520 | FLJ58811 | NLRB1 | psiNAIP | NLR family, apoptosis inhibitory protein | - | HPRD | 11865055 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD | 9556573 |
| NUDT21 | CFIM25 | CPSF5 | DKFZp686H1588 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 | - | HPRD | 14743216 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | - | HPRD,BioGRID | 11585904 |
| PELI3 | MGC35521 | pellino homolog 3 (Drosophila) | - | HPRD | 12874243 |
| PPM1B | MGC21657 | PP2C-beta-X | PP2CB | PP2CBETA | PPC2BETAX | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | - | HPRD,BioGRID | 11104763 |
| PPM1L | MGC132545 | MGC132547 | PP2C-epsilon | PP2CE | PPM1-LIKE | protein phosphatase 1 (formerly 2C)-like | - | HPRD | 12556533 |
| RPLP0 | L10E | MGC111226 | MGC88175 | P0 | PRLP0 | RPP0 | ribosomal protein, large, P0 | - | HPRD | 14743216 |
| RPS3 | FLJ26283 | FLJ27450 | MGC87870 | ribosomal protein S3 | - | HPRD | 14743216 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | - | HPRD | 14743216 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | - | HPRD | 14743216 |
| SMAD6 | HsT17432 | MADH6 | MADH7 | SMAD family member 6 | - | HPRD,BioGRID | 10748100 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD,BioGRID | 12589052 |
| STRADB | ALS2CR2 | CALS-21 | ILPIP | ILPIPA | MGC102916 | PAPK | PRO1038 | STE20-related kinase adaptor beta | - | HPRD,BioGRID | 12048196 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | - | HPRD,BioGRID | 11809792 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Affinity Capture-Western | BioGRID | 14633987 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD | 14743216 |
| TRAF3IP2 | ACT1 | C6orf2 | C6orf4 | C6orf5 | C6orf6 | CIKS | DKFZp586G0522 | MGC3581 | TRAF3 interacting protein 2 | - | HPRD | 12089335 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10094049 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD | 14743216 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | - | HPRD | 14743216 |
| TUBB4 | TUBB5 | beta-5 | tubulin, beta 4 | - | HPRD | 14743216 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | - | HPRD,BioGRID | 12048196 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID TCR JNK PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM TCR SIGNALING | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | 14 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER LARGE VS TINY DN | 45 | 24 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 6Q21 DELETION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 2 UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 455 | 462 | 1A,m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 128 | 134 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-141/200a | 84 | 90 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-143 | 399 | 406 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-339 | 454 | 460 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.