Gene Page: VIPR1
Summary ?
| GeneID | 7433 |
| Symbol | VIPR1 |
| Synonyms | HVR1|II|PACAP-R-2|PACAP-R2|RDC1|V1RG|VAPC1|VIP-R-1|VIPR|VIRG|VPAC1|VPAC1R|VPCAP1R |
| Description | vasoactive intestinal peptide receptor 1 |
| Reference | MIM:192321|HGNC:HGNC:12694|Ensembl:ENSG00000114812|HPRD:01890|Vega:OTTHUMG00000131792 |
| Gene type | protein-coding |
| Map location | 3p22 |
| Pascal p-value | 0.401 |
| Sherlock p-value | 0.814 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.32:CC_BA10_disease_P=0.0371:HBB_BA9_fold_change=-1.32:HBB_BA9_disease_P=0.0105 |
| Fetal beta | -2.786 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0024 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
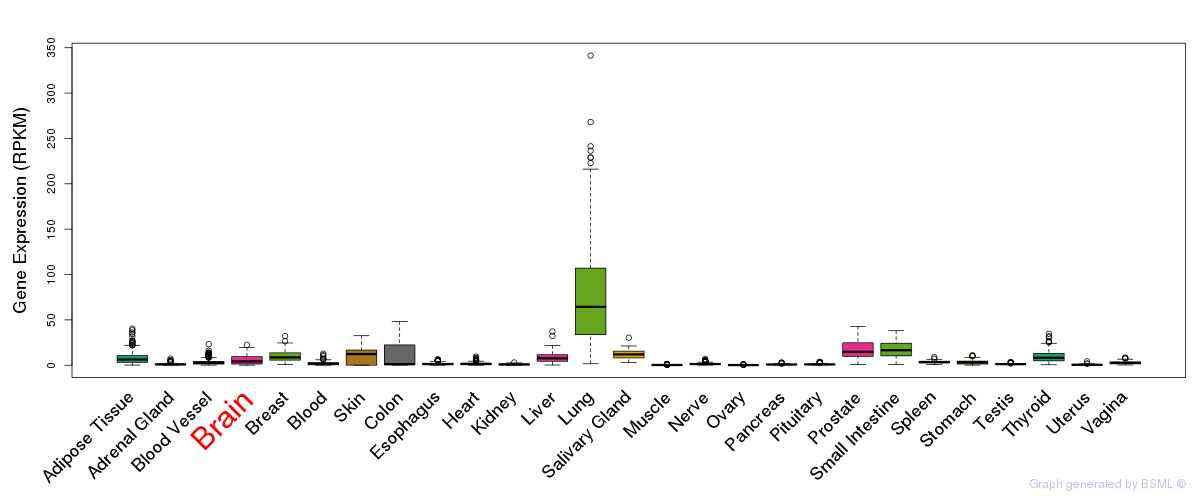
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CBFA2T3 | 0.82 | 0.81 |
| ZNF835 | 0.81 | 0.83 |
| ZNF710 | 0.80 | 0.79 |
| GRK5 | 0.80 | 0.78 |
| NNAT | 0.79 | 0.75 |
| DNMT3A | 0.78 | 0.72 |
| GPC2 | 0.77 | 0.69 |
| ZSWIM5 | 0.77 | 0.79 |
| FUT4 | 0.76 | 0.80 |
| DLL3 | 0.76 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.54 | -0.75 |
| AF347015.31 | -0.54 | -0.74 |
| MT-CO2 | -0.53 | -0.74 |
| AF347015.8 | -0.52 | -0.74 |
| AF347015.33 | -0.52 | -0.70 |
| MT-CYB | -0.52 | -0.72 |
| C5orf53 | -0.52 | -0.61 |
| COPZ2 | -0.51 | -0.64 |
| S100B | -0.50 | -0.67 |
| AF347015.15 | -0.50 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004999 | vasoactive intestinal polypeptide receptor activity | IEA | - | |
| GO:0004999 | vasoactive intestinal polypeptide receptor activity | TAS | 8179610 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004930 | G-protein coupled receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8390245 |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | TAS | 8390245 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008284 | positive regulation of cell proliferation | TAS | 8389448 | |
| GO:0007586 | digestion | TAS | 8390245 | |
| GO:0006955 | immune response | TAS | 8390245 | |
| GO:0006936 | muscle contraction | TAS | 8390245 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8179610 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON TYPE LIGAND RECEPTORS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |