Gene Page: CACNA1A
Summary ?
| GeneID | 773 |
| Symbol | CACNA1A |
| Synonyms | APCA|BI|CACNL1A4|CAV2.1|EA2|FHM|HPCA|MHP|MHP1|SCA6 |
| Description | calcium voltage-gated channel subunit alpha1 A |
| Reference | MIM:601011|HGNC:HGNC:1388|Ensembl:ENSG00000141837|HPRD:03004|Vega:OTTHUMG00000044590 |
| Gene type | protein-coding |
| Map location | 19p13 |
| Pascal p-value | 0.014 |
| Sherlock p-value | 4.203E-5 |
| Fetal beta | -0.694 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CANABINOID DOPAMINE EXCITABILITY SEROTONIN G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 21 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18011401 | 19 | 13617366 | CACNA1A | 5.84E-9 | -0.037 | 3.16E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | CACNA1A | 773 | 0 | trans | ||
| rs2502827 | chr1 | 176044216 | CACNA1A | 773 | 0.01 | trans | ||
| rs12405921 | chr1 | 206695730 | CACNA1A | 773 | 0.12 | trans | ||
| rs17572651 | chr1 | 218943612 | CACNA1A | 773 | 6.714E-4 | trans | ||
| snp_a-1919794 | 0 | CACNA1A | 773 | 0.06 | trans | |||
| rs3811060 | chr2 | 114429210 | CACNA1A | 773 | 0.08 | trans | ||
| rs16829545 | chr2 | 151977407 | CACNA1A | 773 | 3.222E-7 | trans | ||
| rs3845734 | chr2 | 171125572 | CACNA1A | 773 | 1.234E-4 | trans | ||
| rs1967327 | chr2 | 179314358 | CACNA1A | 773 | 0.12 | trans | ||
| rs7584986 | chr2 | 184111432 | CACNA1A | 773 | 9.642E-12 | trans | ||
| rs6729020 | chr2 | 189064719 | CACNA1A | 773 | 0.04 | trans | ||
| rs17762315 | chr5 | 76807576 | CACNA1A | 773 | 0 | trans | ||
| rs7729096 | chr5 | 76835927 | CACNA1A | 773 | 0.04 | trans | ||
| rs1368303 | chr5 | 147672388 | CACNA1A | 773 | 1.101E-7 | trans | ||
| rs6989594 | chr8 | 126303866 | CACNA1A | 773 | 0.08 | trans | ||
| rs17263352 | chr9 | 124811572 | CACNA1A | 773 | 0.17 | trans | ||
| rs1629937 | chr14 | 77059671 | CACNA1A | 773 | 0.04 | trans | ||
| rs17104720 | chr14 | 77127308 | CACNA1A | 773 | 1.567E-4 | trans | ||
| rs17104722 | chr14 | 77138109 | CACNA1A | 773 | 0.14 | trans | ||
| rs17104782 | chr14 | 77156735 | CACNA1A | 773 | 0.14 | trans | ||
| rs6574467 | chr14 | 79179744 | CACNA1A | 773 | 0 | trans | ||
| rs16955618 | chr15 | 29937543 | CACNA1A | 773 | 1.228E-24 | trans | ||
| snp_a-1830894 | 0 | CACNA1A | 773 | 0.01 | trans | |||
| rs16978352 | chr18 | 42944875 | CACNA1A | 773 | 0.19 | trans | ||
| rs17085767 | chr18 | 69839397 | CACNA1A | 773 | 0.15 | trans |
Section II. Transcriptome annotation
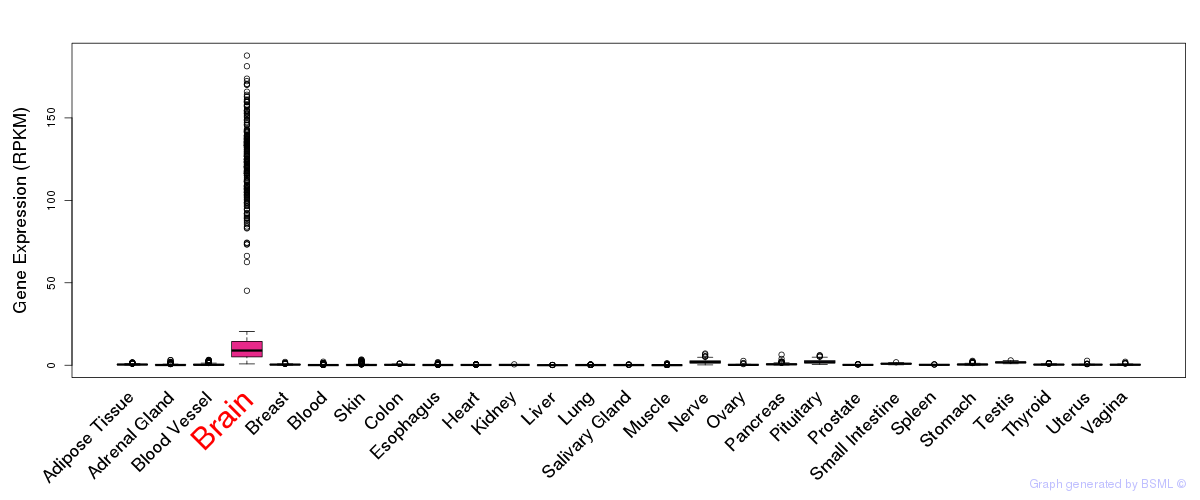
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BAT2L | 0.93 | 0.95 |
| GATAD2B | 0.93 | 0.93 |
| NAV1 | 0.93 | 0.93 |
| KIAA1549 | 0.93 | 0.95 |
| SLC7A6 | 0.93 | 0.93 |
| CELSR3 | 0.93 | 0.95 |
| CRAMP1L | 0.92 | 0.93 |
| MFHAS1 | 0.92 | 0.94 |
| ZMIZ1 | 0.92 | 0.93 |
| RAD54L2 | 0.92 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.68 | -0.77 |
| AF347015.31 | -0.66 | -0.88 |
| S100B | -0.66 | -0.85 |
| AIFM3 | -0.65 | -0.77 |
| C5orf53 | -0.65 | -0.76 |
| FXYD1 | -0.65 | -0.86 |
| MT-CO2 | -0.65 | -0.88 |
| HEPN1 | -0.65 | -0.76 |
| HSD17B14 | -0.65 | -0.79 |
| TSC22D4 | -0.64 | -0.78 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019905 | syntaxin binding | IDA | Synap (GO term level: 5) | 8692999 |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 8692999 |11865310 | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0008331 | high voltage-gated calcium channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007416 | synaptogenesis | IEA | Synap (GO term level: 6) | - |
| GO:0021680 | cerebellar Purkinje cell layer development | IEA | neuron, axon, Synap, dendrite (GO term level: 12) | - |
| GO:0021679 | cerebellar molecular layer development | IEA | neuron, dendrite (GO term level: 12) | - |
| GO:0021702 | cerebellar Purkinje cell differentiation | IEA | neuron, GABA, Brain (GO term level: 15) | - |
| GO:0007274 | neuromuscular synaptic transmission | IEA | neuron, Synap (GO term level: 7) | - |
| GO:0014056 | regulation of acetylcholine secretion | IEA | Neurotransmitter (GO term level: 10) | - |
| GO:0014051 | gamma-aminobutyric acid secretion | IEA | GABA, Brain, Neurotransmitter (GO term level: 10) | - |
| GO:0021590 | cerebellum maturation | IEA | Brain (GO term level: 11) | - |
| GO:0060024 | rhythmic synaptic transmission | IEA | Synap (GO term level: 8) | - |
| GO:0021953 | central nervous system neuron differentiation | IEA | neuron (GO term level: 9) | - |
| GO:0007214 | gamma-aminobutyric acid signaling pathway | IEA | GABA, Neurotransmitter (GO term level: 8) | - |
| GO:0050770 | regulation of axonogenesis | IEA | neuron, axon (GO term level: 13) | - |
| GO:0042133 | neurotransmitter metabolic process | IEA | neuron, axon, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0048791 | calcium ion-dependent exocytosis of neurotransmitter | IEA | Synap, Neurotransmitter (GO term level: 9) | - |
| GO:0035249 | synaptic transmission, glutamatergic | IEA | neuron, glutamate, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0048813 | dendrite morphogenesis | IEA | neurite, dendrite (GO term level: 12) | - |
| GO:0000096 | sulfur amino acid metabolic process | IEA | - | |
| GO:0007204 | elevation of cytosolic calcium ion concentration | ISS | - | |
| GO:0016049 | cell growth | IEA | - | |
| GO:0008219 | cell death | IDA | 16595610 | |
| GO:0006006 | glucose metabolic process | IEA | - | |
| GO:0007628 | adult walking behavior | IEA | - | |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0050885 | neuromuscular process controlling balance | IEA | - | |
| GO:0050883 | musculoskeletal movement, spinal reflex action | IEA | - | |
| GO:0032353 | negative regulation of hormone biosynthetic process | IEA | - | |
| GO:0042403 | thyroid hormone metabolic process | IEA | - | |
| GO:0048266 | behavioral response to pain | IEA | - | |
| GO:0030644 | cellular chloride ion homeostasis | IEA | - | |
| GO:0043113 | receptor clustering | IEA | - | |
| GO:0017158 | regulation of calcium ion-dependent exocytosis | IEA | - | |
| GO:0021750 | vestibular nucleus development | IEA | - | |
| GO:0051899 | membrane depolarization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0042995 | cell projection | IDA | axon (GO term level: 4) | 16595610 |
| GO:0030425 | dendrite | IEA | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0005634 | nucleus | IDA | 16595610 | |
| GO:0005737 | cytoplasm | IDA | 16595610 | |
| GO:0016021 | integral to membrane | TAS | 16595610 | |
| GO:0005886 | plasma membrane | EXP | 8825650 | |
| GO:0005886 | plasma membrane | IDA | 16595610 | |
| GO:0005891 | voltage-gated calcium channel complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TASTE TRANSDUCTION | 52 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION APOBEC2 | 27 | 19 | All SZGR 2.0 genes in this pathway |
| GRASEMANN RETINOBLASTOMA WITH 6P AMPLIFICATION | 14 | 14 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 38 | 44 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.