Gene Page: CALCA
Summary ?
| GeneID | 796 |
| Symbol | CALCA |
| Synonyms | CALC1|CGRP|CGRP-I|CGRP1|CT|KC|PCT |
| Description | calcitonin related polypeptide alpha |
| Reference | MIM:114130|HGNC:HGNC:1437|Ensembl:ENSG00000110680|HPRD:00238|Vega:OTTHUMG00000159731 |
| Gene type | protein-coding |
| Map location | 11p15.2 |
| Pascal p-value | 0.706 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26833169 | 11 | 14994045 | CALCA | 4.48E-5 | -0.381 | 0.021 | DMG:Wockner_2014 |
| cg09188980 | 11 | 14993378 | CALCA | 1E-4 | -0.53 | 0.028 | DMG:Wockner_2014 |
| cg25783352 | 11 | 14993509 | CALCA | 1.029E-4 | -0.449 | 0.028 | DMG:Wockner_2014 |
| cg22863523 | 11 | 14995201 | CALCA | 3.306E-4 | 0.939 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
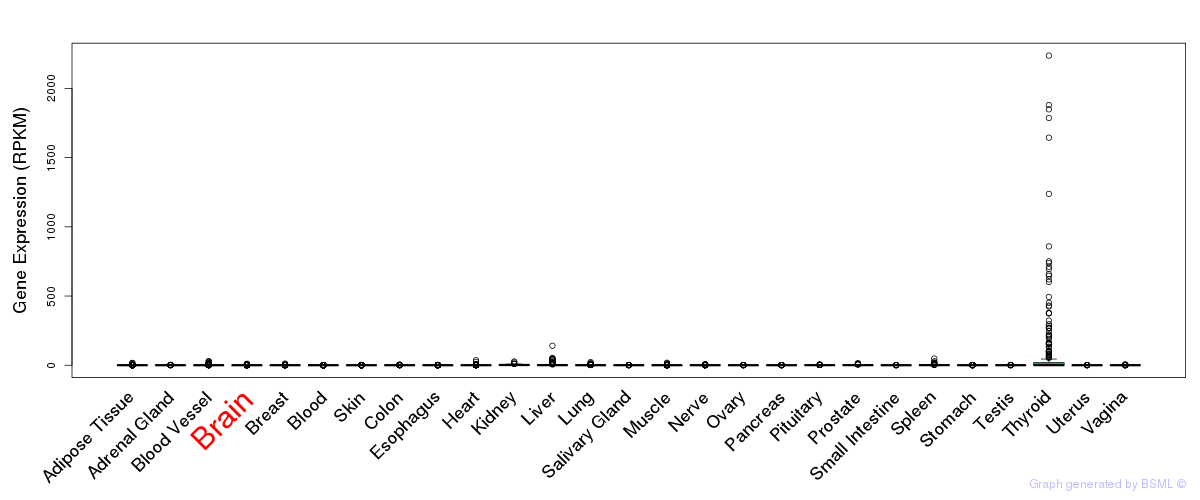
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GOLIM4 | 0.81 | 0.83 |
| COBLL1 | 0.78 | 0.79 |
| TLN1 | 0.75 | 0.81 |
| PTRF | 0.73 | 0.76 |
| CGNL1 | 0.72 | 0.78 |
| FYCO1 | 0.72 | 0.82 |
| COL14A1 | 0.71 | 0.73 |
| GIMAP8 | 0.71 | 0.74 |
| MYH11 | 0.71 | 0.71 |
| FAM129A | 0.70 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF32 | -0.49 | -0.59 |
| COMMD3 | -0.47 | -0.56 |
| NDUFAF2 | -0.47 | -0.55 |
| PFDN5 | -0.47 | -0.59 |
| PHPT1 | -0.46 | -0.57 |
| OXSM | -0.46 | -0.54 |
| C11orf73 | -0.46 | -0.54 |
| MRPS24 | -0.46 | -0.50 |
| AC034193.4 | -0.46 | -0.50 |
| DCTN3 | -0.46 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | IEA | - | |
| GO:0031716 | calcitonin receptor binding | IEA | - | |
| GO:0031716 | calcitonin receptor binding | IPI | 8078488 |8940110 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| GO:0001984 | vasodilation of artery during baroreceptor response to increased systemic arterial blood pressure | IEA | - | |
| GO:0002027 | regulation of heart rate | IEA | - | |
| GO:0001944 | vasculature development | IDA | 17267696 | |
| GO:0001935 | endothelial cell proliferation | IDA | 17267696 | |
| GO:0001976 | regulation of systemic arterial blood pressure by neurological process | IDA | 10642343 | |
| GO:0007190 | activation of adenylate cyclase activity | IDA | 10822112 |16014619 | |
| GO:0007267 | cell-cell signaling | TAS | 2408883 | |
| GO:0009408 | response to heat | IEA | - | |
| GO:0007631 | feeding behavior | IEA | - | |
| GO:0007566 | embryo implantation | IDA | 17983652 | |
| GO:0006954 | inflammatory response | IEA | - | |
| GO:0007159 | leukocyte adhesion | IDA | 1326102 | |
| GO:0016481 | negative regulation of transcription | IDA | 11014233 | |
| GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | IEA | - | |
| GO:0032757 | positive regulation of interleukin-8 production | IDA | 16904178 | |
| GO:0032730 | positive regulation of interleukin-1 alpha production | IDA | 16904178 | |
| GO:0048265 | response to pain | IEA | - | |
| GO:0030279 | negative regulation of ossification | IEA | - | |
| GO:0031645 | negative regulation of neurological system process | IEA | - | |
| GO:0032147 | activation of protein kinase activity | IDA | 17983652 | |
| GO:0045779 | negative regulation of bone resorption | IDA | 10822112 |17241109 | |
| GO:0045778 | positive regulation of ossification | IEA | - | |
| GO:0045651 | positive regulation of macrophage differentiation | IDA | 10822112 | |
| GO:0045762 | positive regulation of adenylate cyclase activity | IDA | 8078488 |11014233 | |
| GO:0045909 | positive regulation of vasodilation | IDA | 3266556 | |
| GO:0045671 | negative regulation of osteoclast differentiation | IDA | 10822112 | |
| GO:0043542 | endothelial cell migration | IDA | 17267696 | |
| GO:0045986 | negative regulation of smooth muscle contraction | IEA | - | |
| GO:0045776 | negative regulation of blood pressure | IDA | 10642343 | |
| GO:0051480 | cytosolic calcium ion homeostasis | IDA | 9685362 | |
| GO:0051482 | elevation of cytosolic calcium ion concentration during G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | IDA | 8078488 |9685362 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0043195 | terminal button | IEA | axon, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0030424 | axon | IEA | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 2408883 |18057382 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 17267696 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT2 TARGETS | 58 | 35 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-488 | 247 | 253 | m8 | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.