Gene Page: HIST1H2BE
Summary ?
| GeneID | 8344 |
| Symbol | HIST1H2BE |
| Synonyms | H2B.h|H2B/h|H2BFH|dJ221C16.8 |
| Description | histone cluster 1, H2be |
| Reference | MIM:602805|HGNC:HGNC:4753|Ensembl:ENSG00000274290|HPRD:11901|Vega:OTTHUMG00000014427 |
| Gene type | protein-coding |
| Map location | 6p22.2 |
| Pascal p-value | 8.884E-4 |
| Fetal beta | 0.504 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIST1H2BE | chr6 | 26184139 | C | A | NM_003523 | p.39S>Y | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11688718 | chr2 | 73760938 | HIST1H2BE | 8344 | 0.12 | trans | ||
| rs4346412 | chr2 | 73843570 | HIST1H2BE | 8344 | 0.19 | trans | ||
| rs4852939 | chr2 | 73856768 | HIST1H2BE | 8344 | 0.18 | trans | ||
| rs10994209 | chr10 | 61877705 | HIST1H2BE | 8344 | 0.09 | trans |
Section II. Transcriptome annotation
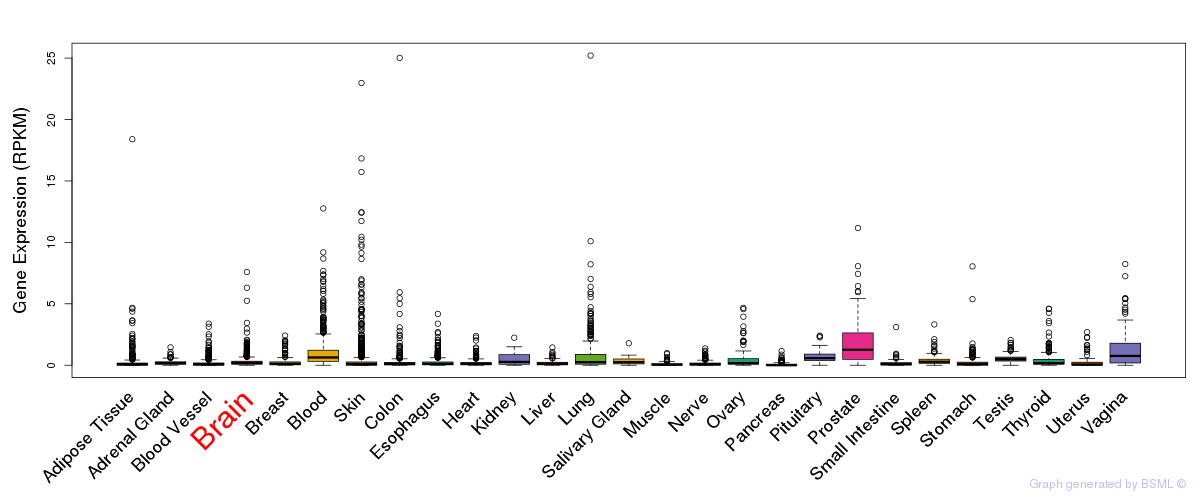
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRUB2 | 0.91 | 0.89 |
| UQCRC1 | 0.91 | 0.92 |
| MRPL38 | 0.91 | 0.88 |
| ANKRD39 | 0.91 | 0.88 |
| WDR45 | 0.90 | 0.90 |
| ERI3 | 0.90 | 0.88 |
| MRPL4 | 0.89 | 0.86 |
| CYC1 | 0.89 | 0.89 |
| NEU1 | 0.89 | 0.87 |
| STOML2 | 0.89 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC010300.1 | -0.53 | -0.67 |
| AF347015.18 | -0.53 | -0.47 |
| EIF5B | -0.49 | -0.57 |
| NSBP1 | -0.48 | -0.49 |
| AL139819.3 | -0.47 | -0.46 |
| AC005921.3 | -0.47 | -0.58 |
| MT-ATP8 | -0.47 | -0.38 |
| AF347015.26 | -0.46 | -0.36 |
| C10orf108 | -0.46 | -0.44 |
| AC016705.1 | -0.45 | -0.44 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I PROMOTER OPENING | 62 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME PACKAGING OF TELOMERE ENDS | 48 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE | 75 | 57 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE UP | 25 | 13 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION UP | 32 | 18 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |