Gene Page: IKBKAP
Summary ?
| GeneID | 8518 |
| Symbol | IKBKAP |
| Synonyms | DYS|ELP1|FD|IKAP|IKI3|TOT1 |
| Description | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| Reference | MIM:603722|HGNC:HGNC:5959|Ensembl:ENSG00000070061|HPRD:04763|Vega:OTTHUMG00000020465 |
| Gene type | protein-coding |
| Map location | 9q31 |
| Pascal p-value | 0.045 |
| Sherlock p-value | 0.766 |
| Fetal beta | 0.532 |
| eGene | Nucleus accumbens basal ganglia |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1609 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
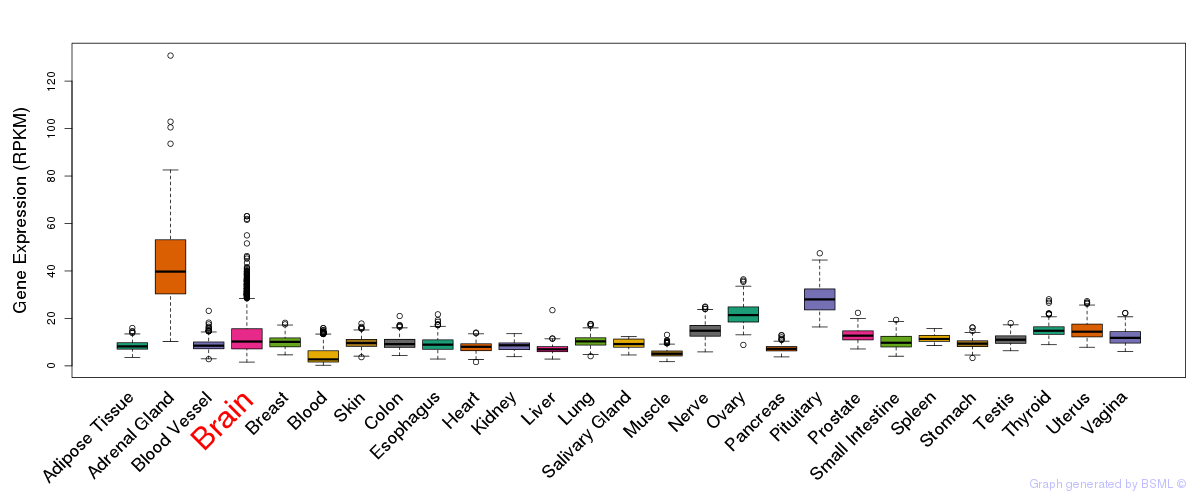
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 9751059 | |
| GO:0003677 | DNA binding | IDA | 11714725 | |
| GO:0005515 | protein binding | IPI | 10094049 |14667819 |15383276 | |
| GO:0008607 | phosphorylase kinase regulator activity | IDA | 11818576 | |
| GO:0016944 | RNA polymerase II transcription elongation factor activity | IDA | 11714725 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | IDA | 11818576 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006461 | protein complex assembly | TAS | 9751059 | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 9751059 | |
| GO:0006955 | immune response | TAS | 9751059 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 11714725 | |
| GO:0005737 | cytoplasm | IDA | 11714725 | |
| GO:0008023 | transcription elongation factor complex | IDA | 11714725 | |
| GO:0016591 | DNA-directed RNA polymerase II, holoenzyme | IDA | 11714725 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Two-hybrid | BioGRID | 16169070 |
| C11orf79 | FLJ20487 | chromosome 11 open reading frame 79 | Two-hybrid | BioGRID | 16169070 |
| CCT5 | CCT-epsilon | CCTE | KIAA0098 | TCP-1-epsilon | chaperonin containing TCP1, subunit 5 (epsilon) | Two-hybrid | BioGRID | 16169070 |
| CDKN2B | CDK4I | INK4B | MTS2 | P15 | TP15 | p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | Two-hybrid | BioGRID | 16169070 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKAP interacts with IKK-alpha. This interaction was modeled on a demonstrated interaction between human IKAP and IKK-alpha from an unspecified species. | BIND | 9751059 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 9751059 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Two-hybrid | BioGRID | 16169070 |
| ELP2 | FLJ10879 | SHINC-2 | STATIP1 | StIP | elongation protein 2 homolog (S. cerevisiae) | Co-purification | BioGRID | 11714725 |
| ELP3 | FLJ10422 | KAT9 | elongation protein 3 homolog (S. cerevisiae) | - | HPRD | 10936 |
| ELP3 | FLJ10422 | KAT9 | elongation protein 3 homolog (S. cerevisiae) | Affinity Capture-Western Co-purification | BioGRID | 11714725 |
| ELP4 | C11orf19 | FLJ20498 | PAX6NEB | PAXNEB | dJ68P15A.1 | elongation protein 4 homolog (S. cerevisiae) | Co-purification | BioGRID | 11714725 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | IKAP interacts with IKK-beta. | BIND | 9751059 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD,BioGRID | 9751059 |
| MAN2A2 | MANA2X | mannosidase, alpha, class 2A, member 2 | Two-hybrid | BioGRID | 16169070 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 9751059 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | IKAP interacts with NIK. | BIND | 9751059 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 12058026 |
| MRPL42 | HSPC204 | MRP-L31 | MRPL31 | MRPS32 | PTD007 | RPML31 | mitochondrial ribosomal protein L42 | Two-hybrid | BioGRID | 16169070 |
| NDUFB9 | B22 | DKFZp566O173 | FLJ22885 | LYRM3 | UQOR22 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa | Two-hybrid | BioGRID | 16169070 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Reconstituted Complex | BioGRID | 9751059 |
| NIPSNAP3A | DKFZp564D177 | FLJ13953 | HSPC299 | MGC14553 | NIPSNAP4 | TASSC | nipsnap homolog 3A (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| PLP2 | A4 | A4-LSB | MGC126187 | proteolipid protein 2 (colonic epithelium-enriched) | Two-hybrid | BioGRID | 16169070 |
| SNAPIN | SNAPAP | SNAP-associated protein | Two-hybrid | BioGRID | 16169070 |
| TAC3 | NKB | NKNB | PRO1155 | ZNEUROK1 | tachykinin 3 | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| TXNDC9 | APACD | thioredoxin domain containing 9 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CD40 PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR REJECTION DN | 10 | 6 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 313 | 319 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-494 | 299 | 306 | 1A,m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.