Gene Page: CBLN1
Summary ?
| GeneID | 869 |
| Symbol | CBLN1 |
| Synonyms | - |
| Description | cerebellin 1 precursor |
| Reference | MIM:600432|HGNC:HGNC:1543|Ensembl:ENSG00000102924|HPRD:02697|Vega:OTTHUMG00000133148 |
| Gene type | protein-coding |
| Map location | 16q12.1 |
| Pascal p-value | 0.015 |
| Fetal beta | 0.451 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 0 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00049033 | 16 | 49317080 | CBLN1 | 3.22E-5 | -0.389 | 0.019 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-4290143 | 0 | CBLN1 | 869 | 0.01 | trans | |||
| rs17572651 | chr1 | 218943612 | CBLN1 | 869 | 0.17 | trans | ||
| rs16829545 | chr2 | 151977407 | CBLN1 | 869 | 1.077E-21 | trans | ||
| rs3845734 | chr2 | 171125572 | CBLN1 | 869 | 4.526E-7 | trans | ||
| rs1967327 | chr2 | 179314358 | CBLN1 | 869 | 0.16 | trans | ||
| rs7584986 | chr2 | 184111432 | CBLN1 | 869 | 2.669E-12 | trans | ||
| rs2183142 | chr4 | 159232695 | CBLN1 | 869 | 3.596E-5 | trans | ||
| rs170776 | chr4 | 173276735 | CBLN1 | 869 | 0.02 | trans | ||
| rs1396222 | chr4 | 173279496 | CBLN1 | 869 | 0.01 | trans | ||
| rs335993 | chr4 | 173321789 | CBLN1 | 869 | 0.05 | trans | ||
| rs335980 | chr4 | 173329784 | CBLN1 | 869 | 0 | trans | ||
| rs335982 | chr4 | 173330945 | CBLN1 | 869 | 0.02 | trans | ||
| rs336016 | chr4 | 173366576 | CBLN1 | 869 | 0.19 | trans | ||
| rs337984 | chr4 | 173411662 | CBLN1 | 869 | 0.01 | trans | ||
| rs17762315 | chr5 | 76807576 | CBLN1 | 869 | 8.137E-4 | trans | ||
| rs10491487 | chr5 | 80323367 | CBLN1 | 869 | 0.14 | trans | ||
| snp_a-4218600 | 0 | CBLN1 | 869 | 0.03 | trans | |||
| rs1380396 | chr5 | 100737044 | CBLN1 | 869 | 0.02 | trans | ||
| rs17136514 | chr5 | 100847200 | CBLN1 | 869 | 0.11 | trans | ||
| rs9968624 | chr5 | 100848223 | CBLN1 | 869 | 0.11 | trans | ||
| rs12196880 | chr6 | 24313746 | CBLN1 | 869 | 0.06 | trans | ||
| rs10806988 | chr6 | 24341768 | CBLN1 | 869 | 0.04 | trans | ||
| rs9461864 | chr6 | 33481468 | CBLN1 | 869 | 3.54E-4 | trans | ||
| rs16890367 | chr6 | 38078448 | CBLN1 | 869 | 0.13 | trans | ||
| rs2961531 | chr7 | 79260819 | CBLN1 | 869 | 0.19 | trans | ||
| rs7787830 | chr7 | 98797019 | CBLN1 | 869 | 0.2 | trans | ||
| rs3118341 | chr9 | 25185518 | CBLN1 | 869 | 0 | trans | ||
| rs1126130 | chr9 | 25209346 | CBLN1 | 869 | 0.13 | trans | ||
| rs1998746 | chr9 | 25211761 | CBLN1 | 869 | 0.13 | trans | ||
| rs9406868 | chr9 | 25223372 | CBLN1 | 869 | 0.01 | trans | ||
| rs2225105 | chr9 | 25480908 | CBLN1 | 869 | 0.01 | trans | ||
| rs11139334 | chr9 | 84209393 | CBLN1 | 869 | 6.463E-4 | trans | ||
| rs2393316 | chr10 | 59333070 | CBLN1 | 869 | 0.12 | trans | ||
| rs9989228 | chr14 | 37809252 | CBLN1 | 869 | 0.17 | trans | ||
| rs16955618 | chr15 | 29937543 | CBLN1 | 869 | 1.787E-32 | trans | ||
| rs2077735 | chr15 | 58479024 | CBLN1 | 869 | 0.07 | trans | ||
| rs9941296 | chr16 | 2759498 | CBLN1 | 869 | 0.09 | trans | ||
| rs4782029 | chr16 | 17248015 | CBLN1 | 869 | 0.17 | trans | ||
| rs16945437 | chr17 | 11797650 | CBLN1 | 869 | 0 | trans | ||
| rs11873184 | chr18 | 1584081 | CBLN1 | 869 | 0.01 | trans | ||
| rs11882889 | chr19 | 19887684 | CBLN1 | 869 | 0 | trans | ||
| rs7274477 | chr20 | 1676942 | CBLN1 | 869 | 0.14 | trans | ||
| rs1041786 | chr21 | 22617710 | CBLN1 | 869 | 4.159E-7 | trans | ||
| rs16981387 | chr22 | 26495864 | CBLN1 | 869 | 0.05 | trans | ||
| rs16986332 | chrX | 10958354 | CBLN1 | 869 | 0 | trans |
Section II. Transcriptome annotation
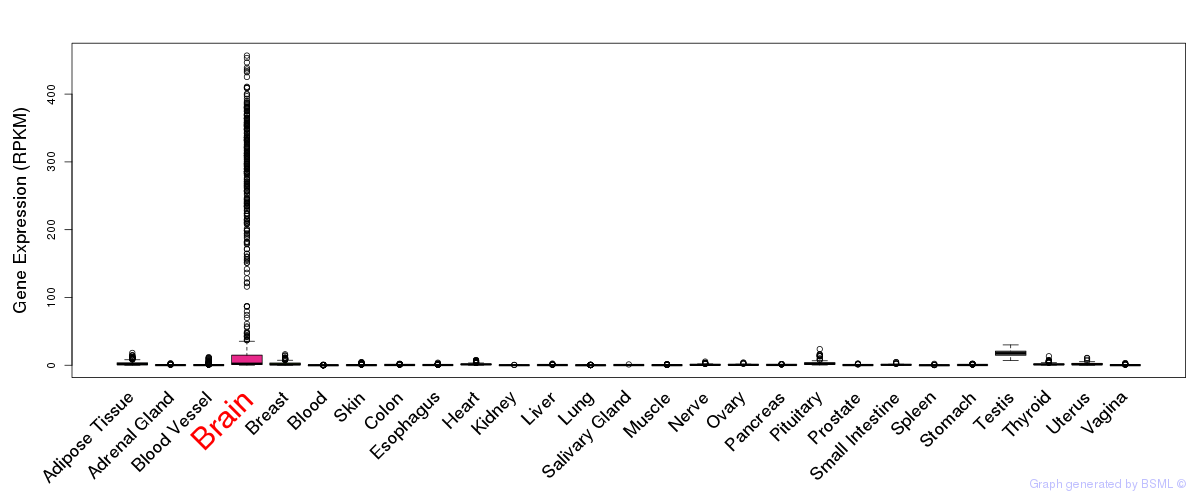
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAL | 0.90 | 0.90 |
| CD9 | 0.90 | 0.90 |
| NINJ2 | 0.89 | 0.87 |
| APOD | 0.89 | 0.88 |
| DBNDD2 | 0.89 | 0.90 |
| PLLP | 0.88 | 0.89 |
| EVI2A | 0.88 | 0.89 |
| RTKN | 0.88 | 0.90 |
| CYP27A1 | 0.87 | 0.89 |
| CRYAB | 0.87 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.61 | -0.62 |
| KIAA1949 | -0.61 | -0.66 |
| CRMP1 | -0.59 | -0.62 |
| ZNF551 | -0.59 | -0.65 |
| AC004017.1 | -0.59 | -0.65 |
| ZBTB8A | -0.59 | -0.62 |
| KIF21B | -0.59 | -0.65 |
| DPYSL3 | -0.59 | -0.63 |
| KIAA1211 | -0.59 | -0.66 |
| GMIP | -0.59 | -0.62 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 1704129 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 1704129 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER UP | 63 | 44 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |