Gene Page: CBR1
Summary ?
| GeneID | 873 |
| Symbol | CBR1 |
| Synonyms | CBR|SDR21C1|hCBR1 |
| Description | carbonyl reductase 1 |
| Reference | MIM:114830|HGNC:HGNC:1548|Ensembl:ENSG00000159228|HPRD:00267|Vega:OTTHUMG00000086618 |
| Gene type | protein-coding |
| Map location | 21q22.13 |
| Pascal p-value | 0.039 |
| Sherlock p-value | 0.308 |
| DEG p-value | DEG:Zhao_2015:p=2.25e-04:q=0.0860 |
| Fetal beta | -1.841 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Nucleus accumbens basal ganglia Myers' cis & trans Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0044 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16906244 | chr8 | 137720394 | CBR1 | 873 | 0.16 | trans | ||
| rs3911702 | chr8 | 137751039 | CBR1 | 873 | 0.08 | trans | ||
| rs7842167 | chr8 | 137841733 | CBR1 | 873 | 0.07 | trans | ||
| rs2845741 | 21 | 37365053 | CBR1 | ENSG00000159228.8 | 1.788E-6 | 0.01 | -77186 | gtex_brain_ba24 |
| rs2845742 | 21 | 37365056 | CBR1 | ENSG00000159228.8 | 1.788E-6 | 0.01 | -77183 | gtex_brain_ba24 |
| rs112953000 | 21 | 37365315 | CBR1 | ENSG00000159228.8 | 9.587E-7 | 0.01 | -76924 | gtex_brain_ba24 |
| rs77372912 | 21 | 37366070 | CBR1 | ENSG00000159228.8 | 1.572E-6 | 0.01 | -76169 | gtex_brain_ba24 |
| rs7280453 | 21 | 37373027 | CBR1 | ENSG00000159228.8 | 6.781E-7 | 0.01 | -69212 | gtex_brain_ba24 |
| rs7280598 | 21 | 37373146 | CBR1 | ENSG00000159228.8 | 6.663E-7 | 0.01 | -69093 | gtex_brain_ba24 |
| rs2032319 | 21 | 37373722 | CBR1 | ENSG00000159228.8 | 6.166E-7 | 0.01 | -68517 | gtex_brain_ba24 |
| rs2835228 | 21 | 37375643 | CBR1 | ENSG00000159228.8 | 2.539E-7 | 0.01 | -66596 | gtex_brain_ba24 |
| rs7282699 | 21 | 37376765 | CBR1 | ENSG00000159228.8 | 4.134E-7 | 0.01 | -65474 | gtex_brain_ba24 |
| rs2276228 | 21 | 37377890 | CBR1 | ENSG00000159228.8 | 4.153E-7 | 0.01 | -64349 | gtex_brain_ba24 |
| rs2254446 | 21 | 37378153 | CBR1 | ENSG00000159228.8 | 4.081E-7 | 0.01 | -64086 | gtex_brain_ba24 |
| rs2013780 | 21 | 37378434 | CBR1 | ENSG00000159228.8 | 3.799E-7 | 0.01 | -63805 | gtex_brain_ba24 |
| rs6517324 | 21 | 37398616 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -43623 | gtex_brain_ba24 |
| rs13047144 | 21 | 37399433 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -42806 | gtex_brain_ba24 |
| rs8132145 | 21 | 37401581 | CBR1 | ENSG00000159228.8 | 4.304E-7 | 0.01 | -40658 | gtex_brain_ba24 |
| rs4817762 | 21 | 37405265 | CBR1 | ENSG00000159228.8 | 4.304E-7 | 0.01 | -36974 | gtex_brain_ba24 |
| rs2835237 | 21 | 37406085 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -36154 | gtex_brain_ba24 |
| rs743422 | 21 | 37406678 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -35561 | gtex_brain_ba24 |
| rs762364 | 21 | 37406849 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -35390 | gtex_brain_ba24 |
| rs72138966 | 21 | 37408608 | CBR1 | ENSG00000159228.8 | 1.74E-6 | 0.01 | -33631 | gtex_brain_ba24 |
| rs2835248 | 21 | 37417489 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -24750 | gtex_brain_ba24 |
| rs2835250 | 21 | 37419207 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -23032 | gtex_brain_ba24 |
| rs2835254 | 21 | 37419786 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -22453 | gtex_brain_ba24 |
| rs2835255 | 21 | 37419844 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -22395 | gtex_brain_ba24 |
| rs1811076 | 21 | 37420102 | CBR1 | ENSG00000159228.8 | 1.83E-7 | 0.01 | -22137 | gtex_brain_ba24 |
| rs2018721 | 21 | 37422966 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -19273 | gtex_brain_ba24 |
| rs762360 | 21 | 37425955 | CBR1 | ENSG00000159228.8 | 8.537E-7 | 0.01 | -16284 | gtex_brain_ba24 |
| rs762361 | 21 | 37425971 | CBR1 | ENSG00000159228.8 | 8.537E-7 | 0.01 | -16268 | gtex_brain_ba24 |
| rs11701298 | 21 | 37431719 | CBR1 | ENSG00000159228.8 | 4.326E-7 | 0.01 | -10520 | gtex_brain_ba24 |
| rs7281482 | 21 | 37433156 | CBR1 | ENSG00000159228.8 | 4.328E-7 | 0.01 | -9083 | gtex_brain_ba24 |
| rs8133454 | 21 | 37437063 | CBR1 | ENSG00000159228.8 | 1.78E-6 | 0.01 | -5176 | gtex_brain_ba24 |
| rs8134917 | 21 | 37437249 | CBR1 | ENSG00000159228.8 | 1.07E-6 | 0.01 | -4990 | gtex_brain_ba24 |
| rs8130498 | 21 | 37438282 | CBR1 | ENSG00000159228.8 | 1.026E-6 | 0.01 | -3957 | gtex_brain_ba24 |
| rs6517327 | 21 | 37440892 | CBR1 | ENSG00000159228.8 | 1.352E-6 | 0.01 | -1347 | gtex_brain_ba24 |
| rs3787728 | 21 | 37443893 | CBR1 | ENSG00000159228.8 | 1.787E-6 | 0.01 | 1654 | gtex_brain_ba24 |
| rs4239798 | 21 | 37446452 | CBR1 | ENSG00000159228.8 | 8.961E-7 | 0.01 | 4213 | gtex_brain_ba24 |
| rs2156407 | 21 | 37447711 | CBR1 | ENSG00000159228.8 | 8.961E-7 | 0.01 | 5472 | gtex_brain_ba24 |
Section II. Transcriptome annotation
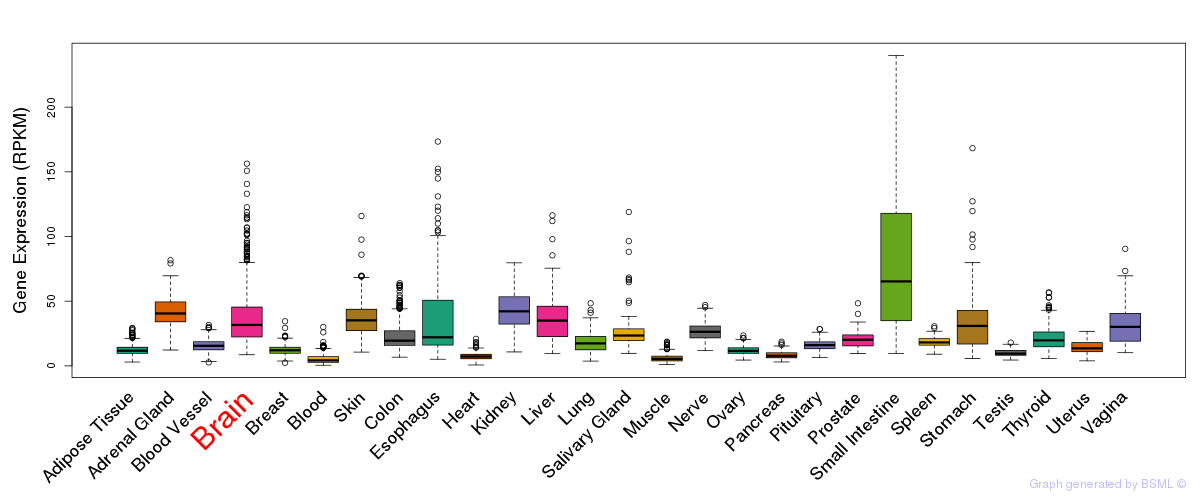
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004090 | carbonyl reductase (NADPH) activity | TAS | 9740676 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0050221 | prostaglandin-E2 9-reductase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0047021 | 15-hydroxyprostaglandin dehydrogenase (NADP+) activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARACHIDONIC ACID METABOLISM | 58 | 36 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MCGARVEY SILENCED BY METHYLATION IN COLON CANCER | 42 | 29 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 UP | 65 | 43 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| WANG ADIPOGENIC GENES REPRESSED BY SIRT1 | 28 | 21 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |